Abstract
Ornamental tobacco (Nicotiana langsdorffii X N. sanderae) secretes a limited array of proteins (nectarins) into its floral nectar. Careful sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of tobacco nectar revealed that a broad protein band from 61 to 65 kD actually consists of five discrete protein bands. N-terminal sequencing and tryptic peptide mass spectrometry fingerprint analysis demonstrated that the upper three bands are isoforms of the same protein, NEC5 (Nectarin V), whereas the lower two bands, NEC4 (Nectarin IV), are related to each other but not to NEC5. Reverse transcription-polymerase chain reaction (RT-PCR) based upon N-terminal sequence of NEC5 generated a short cDNA that encoded the N terminus of the NEC5 protein. Two rounds of inverse-PCR using genomic DNA permitted the isolation of approximately one-half of the coding region of the nec5 gene along with 787 nucleotides of the 5′-flanking region. This DNA fragment was used as a probe to isolate a near full-length nec5 clone from a nectary-derived cDNA library. BLAST analysis identified the nec5 cDNA as a berberine bridge enzyme-like protein. Approximately 40% of the cDNA sequence corresponded to peptides that were identified by tryptic peptide mass spectrometry fingerprint analysis of the NEC5 protein, thereby confirming that this cDNA encoded the NEC5 protein. In-gel assays also demonstrated that NEC5 contains a covalently linked flavin, and it possesses glucose oxidase activity. RT-PCR-based expression analyses showed that nec5 expression is limited exclusively to the nectary gland during late stages of floral development.
During evolution, many angiosperms have developed a remarkable reproductive strategy that relies on animal pollinators to transfer pollen between plants. These plants attract pollinators with floral rewards that encourage insect, avian, and mammalian visitors. Perhaps the primary reward for many species that visit angiosperms is floral nectar. Floral nectar is produced from a terminally differentiated organ frequently located at the base of the flower. Nectar is principally an aqueous carbohydrate solution, containing most frequently sucrose, glucose, and fructose. However, nectars are also known to contain amino acids (Baker and Baker, 1973), organic acids (Baker and Baker, 1975), vitamins (Griebel and Hess, 1940), oils (Vogel, 1969), biological cations (Heinrich, 1989), and proteins (Carter et al., 1999; Carter and Thornburg, 2000, 2003; Thornburg et al., 2003).
Over the past 5 years, our work has demonstrated the presence of a limited array of five proteins, termed nectarins, that are secreted into the nectar of ornamental tobacco (N. langsdorffii X N. sanderae) (Carter et al., 1999). We have demonstrated that the major protein, NEC1 (Nectarin I), is a novel germin-like protein (Carter et al., 1999) that functions as a superoxide dismutase, producing high levels (up to 4 mm) of hydrogen peroxide (Carter and Thornburg, 2000). These levels are antimicrobial (C. Carter and R.W. Thornburg, unpublished data) and have led to the hypothesis that a heretofore unrecognized function of the nectary gland is a defense of the gynoecium from microbial attack (Thornburg et al., 2003). The expression of the nectarins is also tightly regulated. In the case of Nectarin I, the protein is expressed uniquely in the nectary gland and only at times when nectar is actively being secreted from the nectary gland (Carter et al., 1999; Carter and Thornburg, 2003). Furthermore, recent analyses of the nec1 promoter indicate that multiple elements are required to achieve this nectary-specific pattern of expression (Carter and Thornburg, 2003).
The identification of the other nectar proteins has remained elusive until recently. NEC2 (Nectarin II) has been identified as a breakdown product of NEC3 (Nectarin III), which is a dioscorin-like, multifunctional protein with carbonic anhydrase and monodehydroascorbate reductase activities (C. Carter and R.W. Thornburg, manuscript in preparation). Here, we describe NEC5 (Nectarin 5) as flavin-containing berberine bridge enzyme (BBE)-like protein. Although plant nectar has been considered by many to be a simple sugar solution, our work has demonstrated that nectar is a complex biological fluid containing a significant biochemistry.
RESULTS
Attempts to identify and characterize the tobacco nectar proteins were undertaken to evaluate their possible functions within nectar. Our strategy to characterize these proteins was to isolate and subject them to N-terminal sequencing analyses. Based upon N-terminal sequences, efforts were initiated to isolate a corresponding gene for further analysis.
Identification of NEC5 as a BBE-Like Protein
N-Terminal Sequencing of NEC5
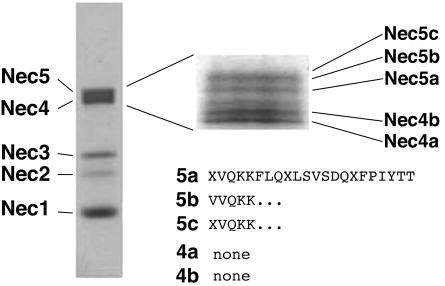
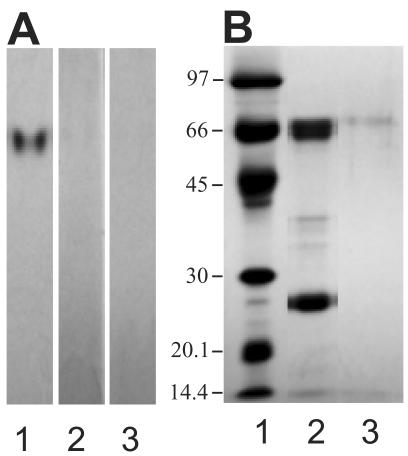
Nectar proteins were separated on an 18-cm, 8% SDS-PAGE and stained with Coomassie Blue R-250. Figure 1 demonstrates that the broad protein band that runs at 61 to 65 kD, formerly termed Nectarin IV (Carter et al., 1999), actually consists of five distinct protein bands. A separate identical gel was blotted onto polyvinyl difluoride (PVDF), and each of these protein bands was excised and subjected to N-terminal sequencing at the Iowa State University Protein Facility (Ames). This analysis revealed that the top three protein bands (NEC5a, 5b, 5c) share the same N-terminal sequence (see Fig. 1). The composite sequence of the upper three bands was determined to be XVQKKFLQXLSVSDQXFPIYTT, where X is unknown. BLAST analysis of this N-terminal sequence failed to identify any potential homologs. The N-terminal amino acid sequencing of the two lowest molecular mass proteins (Nectarin IVa and IVb) of this complex yielded no useful sequence information, and we concluded that these N termini are likely blocked. At this point, these protein bands were renamed NEC4a, 4b, NEC5a, 5b, and 5c in order of increasing molecular mass (see Fig. 1).
Figure 1.
SDS-PAGE analysis of Nectarin 4/5 complex. One hundred microliters of crude nectar was electrophoresed on a 12% Laemmli gel and stained with Coomassie Blue. The inset shows a similar amount of crude nectar run for 12 h on an 18-cm 8% Laemmli gel. The names of the individual proteins are indicated. The N-terminal sequences of each of the peptides are also presented.
Isolation and Characterization of the NEC5 cDNA
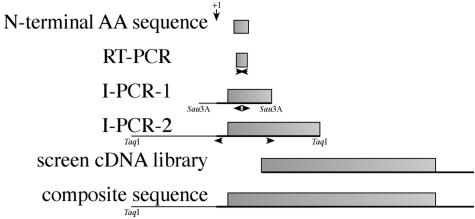
Based upon the N-terminal sequence of the NEC5 proteins, degenerate oligonucleotides for reverse transcription-polymerase chain reaction (RT-PCR) amplification were designed, and a strategy was established for the isolation of the NEC5 coding sequence (Fig. 2). Sequences of the degenerate oligonucleotides are shown in Table I. Total RNA was isolated from mature (Stage 12) nectaries and reverse transcribed using oligo(dT). After first strand synthesis, PCR was used to generate a partial cDNA. This fragment was blunt-end cloned into the HincII site of pUC8 to generate clone pRT507. This partial cDNA was 65 nucleotides in length and matched the first 22 amino acids of the mature protein. The sequence of pRT507 is shown in Figure 3A. The translated amino acid sequence of this clone is compared with the N-terminal sequence of NEC5c.
Figure 2.
Strategy for the isolation of the nec5 gene and cDNA. The N-terminal amino acid sequence permitted the preparation of degenerate primers used for production of a small cDNA, pRT507. Sequence of pRT507 permitted the preparation of specific primers for two rounds of inverse PCR. The insert of pRT516 was used to screen a nectary cDNA library.
Table I.
Oligonucleotides used in this study
| Oligo Name | Sequencea | Purpose |
|---|---|---|
| Nec5-5′ | GCN-GTN-CAR-AAR-AAR-TT | Degenerate RT-PCR |
| Nec5-3′ | GTN-GTR-TAD-ATN-GGR-AA | Degenerate RT-PCR |
| NecV-INV1 | TTA-GTG-ACC-AAA-AAT-TCC-CC | First round inverse-PCR |
| NecV-INV 2 | CAG-ATA-AGC-ATT-GAA-GGA-AC | First round inverse-PCR |
| NecV-FOR1 | CAA-ATG-GGA-CGG-GAA-AAA-TGT-G | Second round inverse-PCR |
| NecV-REV1 | GAT-CCT-CTG-ATA-CAT-AAG-AAA-GGC | Second round inverse-PCR |
| N5-FOR2 | CTC-AGT-ACA-ACA-ACT-AGA-AGG | Internal Primer for RT-PCR |
| N5-REV | GAG-GAA-TAT-AGA-GAC-GAA-TG | Internal primer for RT-PCR |
n = G + A + T + C; R = G + A; D = A + T + C.
Figure 3.
Sequences of nec5. A, Sequence of the pRT507 is presented, along with the sequence of the degenerate oligonucleotides (Nec5-5′ and Nec5-3′), the translation of the pRT507 insert, and the N-terminal amino acid sequence. B, Derived amino acid sequence of the composite NEC5 protein along with the mature N-terminal sequence are presented. The shaded residues indicate the sequences of peptides that correspond to the trypsin fingerprint MS-identified peptides (Table II). The sites of amino acid modification are also indicated. Dots, Sites of N-linked glycosylation; triangle, site of flavin modification. C, Conservation of the predicted flavin-binding region between Nectarin 5 and the EcBBE.
Attempts to use this fragment as a probe to screen a Nicotiana plumbaginifolia genomic library were unsuccessful. Therefore, we performed inverse PCR using oligonucleotides (NecV-INV1 and NecV-INV2 see Table I) that were designed from the internal sequence of the partial cDNA. Genomic DNA, isolated from ornamental tobacco, was subsequently digested with Sau3A I and self-ligated using T4 DNA ligase. Inverse-PCR yielded a 538-nucleotide fragment. This fragment was completely sequenced, and a new set of primers, NecV-FOR1 and NecV-REV1 (Table I), was designed. A second round of inverse-PCR was carried out after digestion of genomic DNA with TaqI and self-ligation. The second round of inverse-PCR resulted in a 1.6-kb fragment that was blunt-end cloned into the HincII site of pUC119 to generate the clone pRT517. This genomic fragment contained about one half of the NEC5 coding region along with 787 nucleotides of flanking sequencing upstream from the ATG start codon. This sequence was deposited into GenBank (accession no. AF503442). Comparison of the amino acid sequence encoded by pRT517 with homologous proteins indicated that this genomic fragment contained no introns. The 5′-flanking regions of nec5 were also analyzed for potential transcription start sites using the neural network promoter prediction site (http://www.fruitfly.org/seq-tools/promoter.html). This tool, which correctly predicted the nec1 transcription start site (Carter and Thornburg, 2003), predicts a single site at A-603 as the transcription start site with high probability.
This larger, inverse-PCR fragment was used to screen a cDNA library derived from mature nectary tissue. A single, strongly hybridizing plaque was taken through two rounds of screening. The plasmid was excised from the phage using the BM25.8 strain of Escherichia coli (CLONTECH, Palo Alto, CA). This clone was then designated as pRT522 and was completely sequenced. This cDNA sequence has been deposited in GenBank (accession no. AF503441).
The isolated cDNA lacked approximately 360 nucleotides at the 5′ end including the first 105 amino acids. However, when the inverse-PCR fragments were aligned with the cDNA, the full open reading frame was revealed (see Fig. 3B).
Characterization of the NEC5 Protein
The full-length nec5 gene encodes a protein of 523 amino acids. The PSORT online analysis tool (Nakai and Kanehisa, 1992) predicted a 21-amino acid N-terminal signal peptide that correctly identified the mature N terminus of NEC5. The mature NEC5 protein has a predicted mature mass of 57,188 D and a pI of 6.40. Also, there are five potential sites of N-glycosylation at N49, N64, N127, N257, and N476.
Tryptic Peptide Mass Fingerprint Analysis of the NEC5 Protein
To confirm that the NEC5 gene encoded the observed proteins, we performed tryptic peptide fingerprint mass spectrometry (MS) on each of the NEC5 proteins. From the initial analysis of the NEC5c protein band, we identified 22 peptides that matched the predicted masses derived from the translated sequence of the NEC5 gene. These peptides covered 39.4% of the total amino acid sequence of the mature protein. Figure 3B and Table II show these identified peptides.
Table II.
Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF) peptide analysis of the NEC5 protein
| Peak No.
|
Mass
|
Amino Acid
|
||
|---|---|---|---|---|
| Observed | Predicted | Sequencea | Range | |
| 1 | 1,817.842 | 1,817.896 | TAWVETGSTIGELYYK | 136-151 |
| 2 | 1,668.856b, c | 1,668.870 | NYSSVLQFSIQNLR | 49-62 |
| 3 | 1,600.817 | 1,600.815 | LYAHMEPLVSTSPR | 444-457 |
| 4 | 1,445.685 | 1,445.688 | FEQMIFTPYGGR | 388-399 |
| 5 | 1,376.738 | 1,376.732 | GGGGNTFGLVLAWK | 222-235 |
| 6 | 1,324.636 | 1,324.635 | SMGEDLFWAIR | 211-221 |
| 7 | 1,277.647 | 1,277.648 | YGLAADNVIDAR | 187-198 |
| 8 | 1,175.623 | 1,175.631 | GLPPTLYSEAK | 351-361 |
| 9 | 1,097.613 | 1,097.563 | FPIYTTNNK | 40-48 |
| 10 | 1,057.588 | 1,057.579 | LHQDLYIR | 272-279 |
| 11 | b,d | 1,045.552 | NITINLDDK | 127-135 |
| 12 | b,d | 939.453 | GNTSYAQAK | 475-483 |
| 13 | 933.481 | 933.479 | VDPSNVFR | 504-511 |
| 14 | b,d | 904.473 | TLEQNATK | 253-260 |
| 15 | 897.463 | 897.447 | WQYVSSK | 265-271 |
| 16 | 888.477 | 888.479 | GESLDVLR | 341-348 |
| 17 | 870.444 | 870.447 | AAYINYR | 437-443 |
| 18 | 827.471 | 827.437 | ESSIHVR | 92-98 |
| 19 | 776.378 | 776.372 | LMDANGR | 199-205 |
| 20 | 665.299 | 665.300 | NNFDR | 492-496 |
| 21 | 610.344b,e | 610.320 | FNTTK | 63-67 |
| 22 | 602.387 | 602.366 | VWGIK | 484-488 |
a Predicted sites of N-glycosylation are shown in bold (peptides 2, 11, 12, 14, and 21). b Contains a predicted site of N-glycosylation. c Peptide observed only in NEC5a and NEC5b isoforms. dPeptide not observed in any of the NEC5 isoforms. e Peptide observed only in NEC5a isoform.
The mass peaks of peptides that were predicted to be N-glycosylated were all absent in the fingerprint analysis of the NEC5c isoform. This suggested that all five sites may be glycosylated (see Table II). If we assume an average mass of 1,500 D per site of glycosylation (Jaikaran et al., 1990), then the predicted mass of the NEC5c protein [57.1 + (5 × 1.5) = 64.6 kD] very closely matches the observed mass from SDS-PAGE of 65 kD. The NEC5a and NEC5b proteins contained an additional peptide peak (Peak 21 in Table II) that was not observed in the NEC5c isoform. This peptide corresponds to amino acids F63 to K67, which contains the N-glycosylation site at NN64-64. The identification of this peptide in the NEC5a and 5b isoforms suggests that this position is not glycosylated in these proteins. In addition, the NEC5a isoform also contained a second peptide peak (Peak 2 in Table II) that was not observed in the NEC5b or NEC5c isoforms. This peptide corresponds to amino acids N49 to R62, which contains the N-glycosylation site at N49. The identification of this peptide in the NEC5a isoform suggests that the NEC5a isoform lacks additional glycosylation. Cumulatively, data for the various NEC5 isoforms (5a, 5b, and 5c) indicate that these proteins are encoded by the same gene product, and the observed differences in masses result from differential N-glycosylation. The two protein bands that appear directly below the NEC5 isoforms (NEC4a and 4b) were also subjected to MALDI-TOF MS tryptic fingerprint analysis. The peptides produced from these proteins are similar to each other but are significantly different from those produced from the NEC5 isoforms and could not be correlated with the sequence of the nec5 cDNA. These data coupled with the finding that the N termini of the NEC4 isoforms appear to be blocked suggests that NEC4 isoforms are not related to the NEC5 gene product.
Characterization of NEC5 as a Flavoprotein
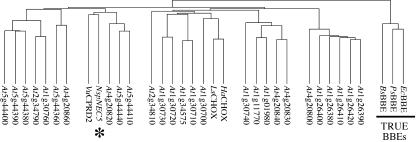
BLAST searches (Altschul et al., 1990) of the full translated NEC5 protein sequence identified it as a BBE-like protein. BBE (reticuline oxidase) was originally identified in California poppy (Eschscholtzia californica; Dittrich and Kutchan, 1991). It catalyzes the conversion of S-reticuline into S-scoulerine and is a major branch point of benzophenanthridine alkaloid biosynthesis in plants (Dittrich and Kutchan, 1991; Bird and Facchini, 2001). A large family of related proteins has been identified in other plants, and in Arabidopsis there are 27 potential homologs (Fig. 4). The three known BBE homologs form an outlying clade in this analysis, and this suggests that the function of most of the BBE-like proteins may not be closely related to the function of these three enzymes. To examine whether NEC5 may be involved in alkaloid biosynthesis in nectar, a Dragendorff test was performed on freshly collected nectar (Baker and Baker, 1982). The negative results indicated that alkaloids are not present in detectable amounts in the nectar of ornamental tobacco and suggest that NEC5 is not likely to be involved in alkaloid biosynthesis in nectar.
Figure 4.
Alignment of BBE-like protein sequences. The tobacco NEC5 is indicated by an asterisk. The three known BBEs form a separate clade and are underlined and indicated as “True BBEs.” The sequences used were: EcBBE, California poppy BBE (GenBank accession no. AF005655); PsBBE, opium poppy (Papaver somniferum) probable reticuline oxidase (AF025430); BsBBE, barberry (Berberis stolonifera) BBE (AF049347); VuCPRD2, cowpea (Vigna unguiculata) drought-induced protein (AB056448); NspNEC5, Nicotiana sp. Nectarin V (AF503441/AF503442); HaCHOX, sunflower (Helianthus annuus) carbohydrate oxidase (AF472609); LsCHOX, lettuce (Lactuca sativa) carbohydrate oxidase (AF472608); and 27 Arabidopsis genes (At1g01980, At1g11770, At1g26380, At1g26390, At1g26400, At1g26410, At1g26420, At1g30700, At1g30710, At1g30720, At1g30730, At1g30740, At1g30760, At1g34575, At2g34790, At2g34810, At4g20800, At4g20820, At4g20830, At4g20840, At4g20860, At5g44360, At5g44380, At5g44390, At5g44400, At5g44410, and At5g44440).
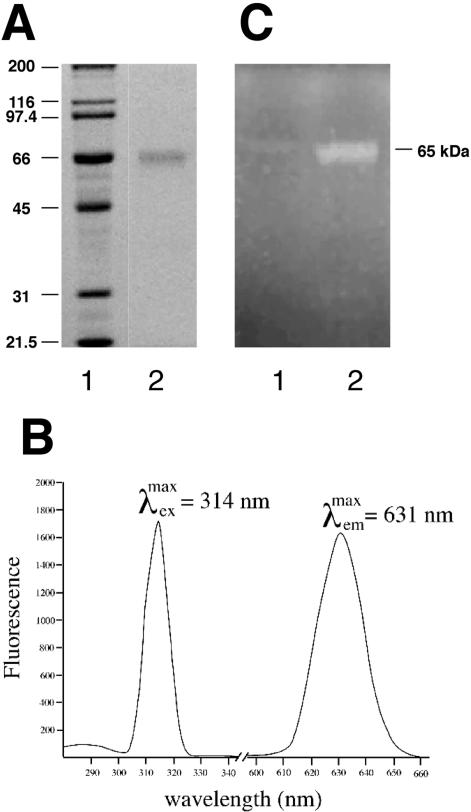
Examination of the best characterized of the BBEs (Kutchan and Dittrich, 1995) revealed that these proteins contain a conserved, covalently bound FAD moiety that is required for activity. The translated NEC5 sequence also contains a homologous consensus flavin-binding domain (Fig. 3C). To test whether NEC5 is a flavoprotein, we purified the protein by ion-exchange chromatography. The presence of a flavin cofactor in the purified NEC5 protein was verified spectroscopically (Fig. 5A). The purified protein's fluorescence spectra were recorded (Fig. 5B). The excitation maximum was found to be 314 nm with a resultant emission maximum at 631 nm. These spectra are consistent with the flavin fluorescence of BBE from California poppy (Kutchan and Dittrich, 1995). To determine whether the cofactor is covalently associated with the protein, both crude nectar and ammonium sulfate precipitated nectar proteins were subjected to standard 12% SDS-PAGE. After electrophoresis, the gel was visualized on a transilluminator at 366 nm. The resultant fluorescence corresponded to the approximately 65-kD NEC5 isoforms (Fig. 5C). These results confirm that NEC5 contains a covalently bound flavin as do the classical BBEs. Further, the tryptic peptide (S101 to R126) that contains the predicted FAD attachment site (H104) is missing from the MALDI-TOF analysis of each of the NEC5 isoforms, suggesting that the mass of this peptide is altered in the mature NEC5 proteins.
Figure 5.
NEC5 is a flavoprotein. A, SDS-PAGE analysis of purified NEC5 protein. Lane 1, Mr standards; lane 2, 15 μg of purified NEC5 protein. B, Fluorescence excitation and emission spectra of the purified NEC5 protein. C, UV fluorescence of NEC5 proteins. Lane 1, One hundred microliters of crude nectar; lane 2, ammonium sulfate precipitated nectar proteins from 1 mL of nectar were electrophoresed on a 12% SDS-PAGE gel and exposed to UV light on a transilluminator (approximately 366 nm), and the illuminated gel was photographed.
Characterization of NEC5 as a Glc Oxidase (GOX)
In addition to characterization of NEC5 as a BBE-like protein, BLAST searches also identified two homologous proteins described simply as carbohydrate oxidases. These proteins were derived from lettuce (AF472608) and sunflower (AF472609). These proteins are labeled LsCHOX and HaCHOX in Figure 4, respectively. Although these proteins are still distantly related to NEC5, the identification of these proteins led us to examine nectar for carbohydrate oxidase activity.
To determine whether nectar contains carbohydrate oxidase activity, crude nectar (25 μL) was applied to an 8% native PAGE and incubated in GOX substrate solution containing 100 mm Glc as described in “Materials and Methods.” This analysis, shown in Figure 6A, revealed a strongly staining band in the presence of glucose (lane 1). Similar experiments were also performed using galactose and mannose as carbohydrate substrates (Lanes 2 and 3). These sugars resulted in no observable activity bands.
Figure 6.
NEC5 has GOX activity. A, Assay for carbohydrate oxidase activity. Twenty-five microliters of crude nectar was electrophoresed on an 8% native page and stained for carbohydrate oxidase activity with 100 mm glucose (lane 1), 100 mm mannose (lane 2), or 100 mm galactose (lane 3). B, Positively staining GOX band processed for Laemmli gel analysis and electrophoresed on a 12% SDS-PAGE stained with Coomassie Blue: lane 1, protein standards; lane 2, 100 μL of crude nectar; lane 3, GOX activity bands excised from native page, applied to SDS-PAGE, and stained with Coomassie Blue.
To verify that this activity is associated with NEC5, the protein bands that positively stained with Glc were excised from the native gel, incubated in SDS-PAGE loading buffer, boiled, applied to standard 12% Laemmli gel, electrophoresed, and stained with Coomassie Blue. The results (Fig. 6B, lane 3) indicated that NEC5 is the only protein observed from the activity band.
Temporal and Spatial Expression of the nec5 Transcript
To evaluate the temporal and spatial patterns of NEC5 expression, RT-PCR was performed. Total RNA was isolated from all major tobacco tissues and from nectaries at various stages of development. This RNA was reverse transcribed using the SMART cDNA synthesis kit (CLONTECH). PCR was then performed using 1 μL of each RT reaction with internal oligonucleotides specific for nec5 (Table I).
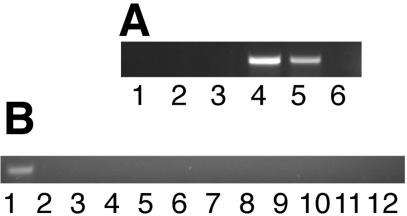
Tobacco floral development can be divided into 12 discreet stages that were defined by Koltunow et al. (1990). During the process of floral development, the nectary also undergoes a series of discrete developmental stages. The initiation of the nectary occurs at a primordial floral stage, when the flower bud is still quite small. By floral Stage 1 the nectary is a well-differentiated ring of cells surrounding the primordial gynoecium. The filling stage of nectary development is characterized by an engorgement of the nectary gland, during which the nectary characteristically enlarges severalfold, beginning at early stages and continuing until through floral Stage 7 (approximately 6 d). Ripening begins about floral Stage 6 and continues through floral Stage 9. During this time frame, the nectary gland turns from green to bright orange because of the synthesis and accumulation of β-carotene (A.B. Barua and R.W. Thornburg, unpublished data). Finally, nectary maturation occurs from floral Stage 9 through floral Stage 12, during which nectar proteins begin to be synthesized. Nectar flow begins at late Stage 10, approximately 12 h before floral opening. Anthesis occurs at Stage 12. The duration of the maturation stage is approximately 12 h. The post-maturation stage comprises all stages that occur after anthesis and lasts as long as the flower is receptive to pollination. During this time, nectar flow continues; however, the flow is stronger at early times and weakens the longer the flower remains open. This stage can sometimes last 6 to 8 d if the flower is not pollinated. Successful pollination leads to fruit development, whereas unpollinated flowers are eventually dropped from the plant. We examined Stage 2 (small floral bud), Stage 6 (immature nectary, green color), Stage 8 (immature nectary, starting to turn yellow/orange), Stage 10 (presecretory), and Stage 12 (mature, secretory) nectaries. Figure 7A demonstrates that the nec5 transcript is not expressed in the immature nectary gland (Stages 2-8) but is found in Stages 10 and 12. After pollination, no nec5 transcript was detectable.
Figure 7.
NEC5 expression patterns. Whole RNA was isolated from the indicated tissues, and RT-PCR was performed as in “Materials and Methods.” A, Temporal expression of nec5 transcript in ornamental tobacco nectaries. The stage of the tissue that was the source of the RNA are: lane 1, stage 2; lane 2, stage 6; lane 3, stage 8; lane 4, stage 10; lane 5, stage 12; lane 6, nectary tissues post-fertilization. B, Spatial expression of nec5 transcript in ornamental tobacco tissues. The floral organ or vegetative tissue that was the source of RNA are: lane 1, nectary; lane 2, ovary; lane 3, style; lane 4, stigma; lane 5, floral tube; lane 6, anther/filament; lane 7, petal; lane 8, sepal; lane 9, receptacle; lane 10, leaf; lane 11, stem; lane 12, root.
To determine spatial expression patterns of NEC5, all major floral and vegetative organs were examined by RT-PCR (Fig. 7B). nec5 transcript expression was limited solely to the nectary tissues. All vegetative organs and all floral organs with the exception of the nectary gland lacked nec5 transcript expression. These studies demonstrated that nec5 is expressed exclusively in the nectary gland. Both the spatial and temporal expression profiles of NEC5 very closely mirror those of NEC1 (Carter et al., 1999; Carter and Thornburg, 2003).
Promoter Comparison Analysis
Because the expression patterns of NEC5 so closely resembled those of NEC1, we next evaluated the 787 nucleotides of nec5 promoter for similarities with the nec1 promoter. To compare these sequences, we initially searched for identity between the two promoters; however, no significant identity between these promoters was found. Recent analysis of the nec1 promoter reveals that multiple DNA elements are involved in generating the nectary-specific phenotype. One of these elements limits expression of the gene in the petals, whereas another element controls the temporal expression (Carter and Thornburg, 2003). This second, temporal regulatory element contains a consensus MYB-binding site. Therefore, we searched the isolated nec5 promoter for consensus MYB-binding sites. This search verified that the nec5 promoter did contain a consensus MYB-binding site (Table III). This site, located at nucleotides 207 to 215 (-395 to -387 relative to the predicted transcription start site) is very similar to MYB-binding sites present in other floral promoters.
Table III.
Conserved MYB sites in nectary expressed promoters
| Nectary Expressed Promoter | GenBank Accession No. | Sequence | Location |
|---|---|---|---|
| myb consensus | NMACCWAMCa | ||
| nec5 | AF503442 | TCACCTAAA | 207 to 215b |
| nec1 | AF132671 | TCACCTAAC | −889 to −891c |
| gPAL2 | d | TCACCTAAC | −238 to −246c |
| Petunia (Petunia hybrida) Necl | AX006361 | TCACCTAAA | 400 to 408b |
| Petunia Necl | AX006361 | TCACCTAAA | 1,621 to 1,629b |
a Where M = A/C and W = A/T. b Relative to the first nucleotide in the GenBank accession. c Relative to the transcription start site. d Not available (Sablowski et al., 1994).
DISCUSSION
We previously described a small set of proteins that are expressed in the nectar of tobacco plants. These proteins are termed nectarins. NEC1, a 29-kD monomer, the most abundant nectar protein, was identified as a manganese-containing, germin-like, superoxide dismutase (Carter et al., 1999; Carter and Thornburg, 2000). The nec1 promoter was later isolated, and reporter constructs demonstrated that the gene is expressed exclusively in actively secreting nectary tissue (Carter and Thornburg, 2003). NEC3 is a dioscorin-like polyfunctional protein that has carbonic anhydrase and monodehydroascorbate reductase activity (C. Carter and R.W. Thornburg, manuscript in preparation). We now have identified the NEC5 protein, determined enzymatic activities, isolated the corresponding cDNA/gene, and resolved its patterns of expression.
N-terminal sequencing of NEC5 isoforms did not reveal any potential homologs, but the N-terminal amino acid sequence permitted us to isolate cDNAs and genomic clones that encode the NEC5 proteins. BLAST analysis of the cloned fragments identified NEC5 as a BBE-like protein. BBE catalyzes the conversion of s-reticuline into s-scoulerine and is a major branch point of benzophenanthridine alkaloid biosynthesis in plants (Dittrich and Kutchan, 1991; Bird and Facchini, 2001). However, most BBE-like proteins have unknown functions. Because the nectar of ornamental tobacco lacks alkaloids, we concluded that NEC5 likely had no role in alkaloid metabolism. Major common characteristics of these BBE-like proteins include putative signal peptides, sites of N-glycosylation, and a conserved flavin-binding domain. The conserved flavin-binding domain in NEC5 is from amino acids 71 to 225, and a covalent site of attachment is predicted at His-104. Fluorometric and in-gel data confirmed the presence of a covalently attached FAD to NEC5.
Previous studies on NEC1 demonstrated that the presence of a signal peptide is likely required for the observed extracellular localization (Carter et al., 1999). N-terminal sequencing suggests that the NEC5 signal peptide that is cleaved between residues D23 and V24.
Flavin-containing proteins generally catalyze redox reactions. The fact that NEC5 contains this cofactor at a conserved site suggested that this protein may also be involved in regulating the oxidative state of nectar. Our analyses indicate that NEC5 contains GOX activity and may function in generating the high levels of hydrogen peroxide found in nectar. GOX catalyzes the oxidation of d-Glc to d-gluconic acid and hydrogen peroxide (Pazur and Kleppe, 1964). The best characterized form of the enzyme, from the fungi Aspergillus niger, is a homodimer, contains two non-covalently bound molecules of FAD, and is highly specific for Glc (Pazur and Kleppe, 1964). The functionally equivalent hexose oxidase (HOX) from the red algae Chondrus crispus can use multiple mono- and disaccharides, lacks FAD, and likely requires copper for activity (Hansen and Stougaard, 1997). Interestingly, although both NEC5 and GOX contain bound flavin, the enzymes share very little identity with one another. Conversely, NEC5 shares a small but significant 23% identity (39% including conserved substitutions) with the copper-containing HOX. Experiments are underway to characterize the absolute requirements for NEC5 GOX activity.
With the high concentration of simple sugars present in tobacco nectar (35% [w/v]), NEC5 GOX activity likely contributes toward the antimicrobial levels of hydrogen peroxide found therein. Interestingly, honey is known for its antimicrobial nature and has been used in wound dressings (Bang et al., 2003). It is thought that GOX is secreted from the hypopharyngeal gland of worker honeybees (Apismellifera) into collected nectar, and studies have demonstrated that the hydrogen peroxide present in honey is at least partly responsible for its antiseptic powers (Bang et al., 2003). Thus, analogous systems may exist in tobacco nectar and in honey for limiting the growth of microorganisms.
Only a handful of BBE genes and their products have been characterized to date (Kutchan and Dittrich, 1995; Facchini et al., 1996; Ikezawa et al., 2003). The “true” BBE genes are involved in the production of benzylisoquinoline alkaloids. Interestingly, there are many BBE-like genes reported in plants that do not contain these alkaloids (Arabidopsis alone contains 27 BBE-like genes). This strongly suggests that the BBE family of proteins is involved in multiple capacities. This is not necessarily a unique finding. For example, the germin gene family in plants is found in large numbers (Arabidopsis has 35 germin-like protein family members and the reported functions of these proteins include oxalate oxidase, superoxide dismutase, auxin-binding, protease inhibitor, and ADP-Glc pyrophosphatase activities (Lane et al., 1993; Ohmiya et al., 1998; Carter and Thornburg, 2000; Rodriguez-Lopez et al., 2001; Segarra et al., 2003). It is likely with time that numerous functions for the BBE gene family will be found. Glucose oxidase appears to be one such function.
The patterns of NEC5 expression are very similar to the patterns of expression of NEC1. This suggests that the promoters regulating these two genes may share common mechanisms regulating their expression. Some floral-specific promoters are known to be regulated by a MYB family member known as MYB305 (Sablowski et al., 1994). Similar MYB sequences are also found in the promoter of the petunia NecI gene (Table III), an unrelated gene, whose product may be involved in nectar secretion (Ge et al., 2000). Finally, PsMYB26, which is a pea (Pisum sativum) MYB305 homolog has been reported to bind in vitro to the sequence TAACCTAAC (Uimari and Strommer, 1997), further suggesting that MYB305 proteins may bind to and regulate the nectarin promoters. Comparison of the nec5 promoter with the nec1 promoter reveals the presence of similar MYB-binding sites that have also been identified in other nectary expressed genes (Table III). Further, the temporal expression pattern is consistent with the expression being driven by a MYB-like factor. MYB proteins are known to be expressed in tobacco flowers at Stage 10 just before anthesis (Sablowski et al., 1994). We recently have isolated several cDNA clones encoding an MYB305-like protein that are expressed in the mature (Stage 12) nectary gland (D.Y. Yin and R.W. Thornburg, unpublished data). We are currently expressing the tobacco MYB305-like protein to determine whether this protein can directly interact with the MYB-binding sites of the nec1 and nec5 promoters.
MATERIALS AND METHODS
Reagents
Reagents and chemicals were obtained from Fisher Scientific (Pittsburgh) or from Sigma (St. Louis) unless otherwise noted.
Plant Material
The line of ornamental tobacco plants used in this study was derived from an interspecific cross between Nicotiana langsdorfii and Nicotiana sanderae. Both of these species are diploid and belong to the Alatae section of Nicotiana. These plants were previously used to study a genetic instability (Kornaga et al., 1997) and the tobacco nectar proteins (Carter et al., 1999; Carter and Thornburg, 2000). Nectar was collected as described (Carter et al., 1999).
N-Terminal Sequencing
Crude nectar protein (100 μg) was subjected to an 8% SDS-PAGE (Laemmli, 1970), briefly stained with Coomassie Blue R-250, and electrophoretically transferred to PVDF membrane. The stained protein bands were excised from the PVDF membrane and sequenced at the Iowa State University Protein Facility on a 477A protein sequencer/120A analyzer (PE-Applied Biosystems, Foster City, CA) using sequential Edman degradation.
MALDI-TOF MS
Crude nectar protein (100 μg) was subjected to an 8% SDS-PAGE and briefly stained with Coomassie Blue G-250. Corresponding bands were excised, digested with trypsin, and analyzed by MALDI-TOF MS (Wang et al., 2000). Masses were collected over a range of 600 to 4,000 D on a TOF mass analyzer (Lasermat 2000 MALDI, Finnigan, Madison, WI).
cDNA Library Construction
Total RNA was isolated from Stage 12 nectaries by the method of Chomczynski and Sacchi (1987), and mRNA was isolated using the PolyATract mRNA isolation kit (Promega, Madison, WI). Purified mRNA was then used to create a cDNA library using the SMART cDNA library construction kit according to the manufacturer's instructions (CLONTECH). The library was packaged using Gigapack Gold III packaging extracts and amplified according to the manufacturer's instructions (Stratagene, La Jolla, CA).
Cloning of the NEC5 cDNA/Gene
Degenerate oligonucleotides, Nec5-5′ and Nec5-3′ (see Table I), derived from the N-terminal amino acid sequence of NEC5 were used for RT-PCR amplification of the nec5 cDNA. Total RNA was isolated (Chomczynski and Sacchi, 1987) and reverse transcribed using oligo(dT) according to standard methods (Ausubel et al., 1992). After first strand synthesis, PCR using the degenerate oligonucleotides was performed according to standard protocols (Erlich, 1989) to generate a partial cDNA. Other DNA methods were performed according to standard protocols (Sambrook et al., 1989; Ausubel et al., 1992). Genomic DNA was isolated from ornamental tobacco leaf tissue according to Ausubel et al. (1992).
RT-PCR Determination of NEC5 Expression Patterns
RNA was isolated from fresh tissues (Chomczynski and Sacchi, 1987), and 1 μg of total RNA was used for first strand cDNA synthesis according to standard methods using oligo(dT) (Ausubel et al., 1992). Internal oligonucleotides, N5-FOR2 and N5-REV1 (see Table I), were designed, and standard RT-PCR was performed. PCR reactions were analyzed by 1% (w/v) Tris-acetic acid-EDTA-agarose electrophoresis.
Carbohydrate Oxidase Assay
After native PAGE electrophoresis, gels were incubated at room temperature overnight in a solution of 50 mm Tris (pH 8.1), 37% (v/v) ethanol containing 0.5 mg mL-1 4-chloro-1-naphthol, 5 units mL-1 horseradish peroxidase, and either 100 mm glucose, mannose, or galactose. Activity appeared as a blue band on a clear background. GOX from Aspergillus niger was used as a positive control.
Purification of NEC5 from Crude Nectar
One milliliter of crude nectar was mixed with 100 μL of 0.1 m sodium phosphate buffer (pH 5.8) and loaded onto a 1-mL CM-Sephadex column (Amersham Biosciences, Piscataway, NJ) pre-equilibrated with 10 mm sodium phosphate buffer (pH 5.8). The column was then washed with five 1-mL volumes of 50 mm sodium phosphate buffer (pH 5.8), and each wash was collected. The protein profile of each fraction was evaluated by SDS-PAGE. Those fractions that contained only NEC5, as determined by SDS-PAGE, were pooled and concentrated to approximately 100 μL with a 10,000 MWC UltraFuge ultrafiltration centrifuge filter (Osmonics, Minnetonka, MN). Because of the limited complexity of the proteins in nectar, this simple procedure permits the separation of NEC5 from the other nectarins.
Acknowledgments
The authors would like to thank Mr. Yingchung Wang and Dr. Parag Chitnis for assistance with the MALDI-TOF MS analyses.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027482.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1992) Short Protocols in Molecular Biology. John Wiley & Sons, New York, p 917
- Baker HG, Baker I (1973) Amino acids in nectar and their evolutionary significance. Nature 241: 543-5454693956 [Google Scholar]
- Baker HG, Baker I (1975) Studies of nectar-constitution and pollinator-plant coevolution. In LE Gilbert, PH Raven, eds, PH Coevolution of Animals and Plants. University of Texas Press, Austin, pp 100-140
- Baker HG, Baker I (1982) Floral nectar sugar constituents in relation to pollinator type. In CE Jones, RJ Little, eds, Handbook of Experimental Pollination Biology. Van Nostrand-Reinhold, New York, pp 107-115
- Bang LM, Buntting C, Molan P (2003) The effect of dilution on the rate of hydrogen peroxide production in honey and its implications for wound healing. J Altem Complement Med 9: 267-273 [DOI] [PubMed] [Google Scholar]
- Bird DA, Facchini PJ (2001) Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta 213: 888-897 [DOI] [PubMed] [Google Scholar]
- Carter C, Graham R, Thornburg RW (1999) Nectarin I is a novel, soluble germin-like protein expressed in the nectar of Nicotiana sp. Plant Mol Biol 41: 207-216 [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW (2000) Tobacco Nectarin I: purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J Biol Chem 275: 36726-36733 [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW (2003) The nectary-specific pattern of gene expression is regulated by multiple promoter elements in the tobacco Nectarin I promoter. Plant Mol Biol 51: 451-457 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156-159 [DOI] [PubMed] [Google Scholar]
- Dittrich H, Kutchan TM (1991) Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc Natl Acad Sci USA 88: 9969-9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich HA (1989) PCR Technology: Principles and Applications for DNA Amplification. p 311
- Facchini PJ, Penzes C, Hohnson AG, Bull D (1996) Moledular characterization of berbenne bridge enzyme gene from opium poppy. Plant Physiol 112: 1669-1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge YX, Angenent GC, Wittich PE, Peters J, Franken J, Busscher M, Zhang LM, Dahlhaus E, Kater MM, Wullems GJ et al. (2000) NEC1, a novel gene, highly expressed in nectary tissue of Petunia hybrida. Plant J 24: 725-734 [DOI] [PubMed] [Google Scholar]
- Griebel C, Hess G (1940) The vitamin C content of flower nectar of certain Labiatae. Zeit Untersuch Lebensmitt 79: 168-171 [Google Scholar]
- Hansen OC, Stougaard P (1997) Hexose oxidase from the red alga Chondrus crispus: purification, molecular cloning, and expression in Pichia pastoris. J Biol Chem 272: 11581-11587 [DOI] [PubMed] [Google Scholar]
- Heinrich G (1989) Analysis of cations in nectars by means of a laser microprobe mass analyser (LAMMA). Beitr Biol Pflanz 64: 293-308 [Google Scholar]
- Ikezana N, Tanaka M, Nagayoshi M, Shinkyo R, Sakaki T, Inouye K, Sato F (2003) Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel p450 family, from cultured coptis japonicus cells. J Biol Chem 278: 38557-38565 [DOI] [PubMed] [Google Scholar]
- Jaikaran ASI, Kennedy TD, Dratewka-Kos E, Lane BG (1990) Covalently bonded and adventitious glycans in germin. J Biol Chem 265: 12503-12512 [PubMed] [Google Scholar]
- Koltunow AM, Truettner, J, Cox KH, Wallroth M, Goldberg RB (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2: 1201-1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornaga T, Zyzak DV, Kintinar A, Baynes J, Thornburg R (1997) Genetic and biochemical characterization of a “lost” unstable flower color phenotype in interspecific crosses of Nicotiana sp. WWW J Biol 2: 8 [Google Scholar]
- Kutchan TM, Dittrich H (1995) Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J Biol Chem 270: 24475-24481 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685 [DOI] [PubMed] [Google Scholar]
- Lane BG, Dunwell JM, Ray JA, Schmitt MR, Cuming AC (1993) Germin, a protein marker of early plant development, is an oxalate oxidase. J Biol Chem 268: 12239-12242 [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14: 897-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya A, Tanaka Y, Kadowaki K, Hayashi T (1998) Cloning of genes encoding auxin-binding proteins (ABP19/20) from peach: significant peptide sequence similarity with germin-like proteins. Plant Cell Physiol 39: 492-499 [DOI] [PubMed] [Google Scholar]
- Pazur JH, Kleppe K (1964) The oxidation of glucose and related compounds by glucose oxidase from Aspergillus niger. Biochemistry 35: 578-583 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez M, Baroja-Fernandez E, Zandueta-Criado A, Moreno- Bruna B, Munoz FJ, Akazawa T, Pozueta-Romero J (2001) Two isoforms of a nucleotide-sugar pyrophosphatase/phosphodiesterase from barley leaves (Hordeum vulgare L.) are distinct oligomers of HvGLP1, a germin-like protein. FEBS Lett 490: 44-48 [DOI] [PubMed] [Google Scholar]
- Sablowski R, Moyano E, Culianez-Macia F, Schuch W, Martin C, Bevan M (1994) A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J 13: 128-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Segarra CI, Casalongue CA, Pinedo ML, Ronchi VP, Conde RD (2003) A germin-like protein of wheat leaf apoplast inhibits serine proteases. J Exp Bot 54: 1335-1341 [DOI] [PubMed] [Google Scholar]
- Thornburg RW, Carter C, Powell A, Rizhsky L, Mittler R, Horner HT (2003) A major function of the tobacco floral nectary is defense against microbial attack. Plant Syst Evol 238: 211-218 [Google Scholar]
- Uimari A, Strommer J (1997) Myb26: a MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J 12: 1273-1284 [DOI] [PubMed] [Google Scholar]
- Vogel S (1969) Flowers offering fatty oil instead of nectar. Abstracts of the XIth International Botanical Congress, Seattle. p 229
- Wang Y, Sun J, Chitnis P (2000) Proteomic study of the peripheral proteins from thylakoid membranes of the cyanobacterium Synechocystis sp. PCC6803. Electrophoresis 21: 1746-1754 [DOI] [PubMed] [Google Scholar]