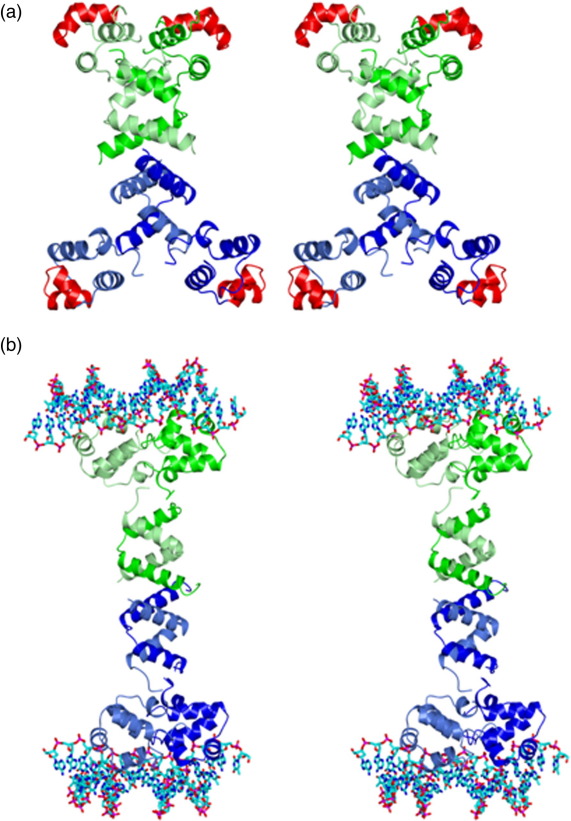
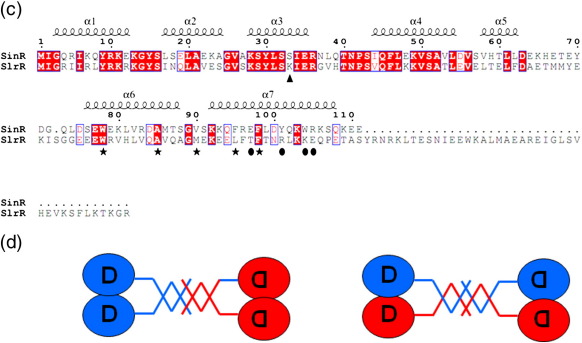
Fig. 5.
The SinR tetramer and SlrR comparison. (a) Stereo model of a SinR tetramer generated by superimposing the coordinates of SinR from the SinR–SinI complex onto each of the four chains of the SinR(74–111) tetramer. The chains of one dimer are coloured light green and green, while those of the other chain are in light blue and blue. The HTH motif is coloured red for all four chains. The exact position of the N-terminal domains in relation to the rest of the protein is expected to be variable due to the flexible linker connecting the domains. (b) Stereo model of a SinR tetramer bound to DNA. Pairs of DNA binding domains from the model generated above were brought together to form dimers matching the molecular packing in the SinR(1–69) crystal. The DNA duplexes are taken from the Cro–DNA (PDB entry 3CRO) complex following least-squares superposition of Cro protein Cα atoms onto the SinR(1–69) dimer. (c) A sequence alignment of SinR and SlrR. Identical residues are highlighted red, with similar residues boxed. Residues of the intermolecular hydrophobic core in SinR are indicated with asterisks, and those involved in dimer–dimer interactions are denoted by filled ovals. The alignment was created using ClustalW and ESPript.17 (d) SinR–SlrR complexes based on a heterodimer of dimers (left) or a dimer of heterodimers (right) models.