Figure 1.
TmMreB Binds and Distorts Lipid Membranes as Shown by Electron Cryomicroscopy
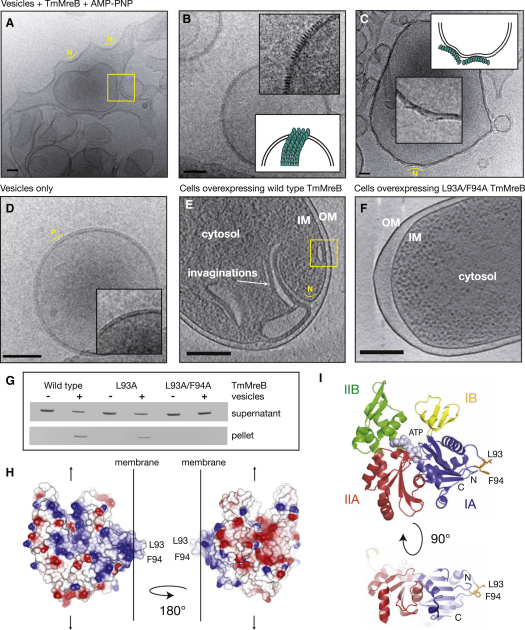
(A–C) Vesicles mixed with purified, untagged Thermotoga maritima (Tm) MreB protein (pFE349) in the presence of AMP-PNP, showing regular protein structures and gross morphological distortions. Schematic insets indicate how TmMreB (cyan) is thought to act on the bilayer. Scale bars, 50 nm.
(D) Negative control showing vesicle only. Scale bar, 50 nm.
(E and F) Section through a 3D electron cryotomography reconstruction of an E. coli cell containing high levels of wild-type, untagged TmMreB (pFE309) (E) or untagged L93A/F94A TmMreB (pJS101) (F). See also Movie S1. Protein expression levels are roughly equal as shown by whole-cell SDS-PAGE analysis in Figure S1B. Inner membrane (IM), outer membrane (OM), cytosol, and membrane invaginations are indicated. Scale bar, 250 nm. Yellow boxes indicate areas where two TmMreB surfaces interact (see also Movie S2), and yellow lines highlight regions of negative (N) and positive (P) curvature.
(G) Vesicle-pelleting assay showing that purified, his-tagged TmMreB (pFE52) directly binds to membranes and that a single (MreB_L93A, pJS104) and double mutation (MreB_L93A/F94A, pJS105) in the membrane insertion loop results in partial and complete loss of membrane binding, respectively.
(H) Schematic showing the known structure of TmMreB and its predicted interaction with the membrane. The residues responsible for membrane binding, L93 and F94, are highlighted, and arrows show the direction of polymerization. The protein surface is colored according to charge, with positive regions colored in blue and negative regions in red.
(I) The crystal structure of TmMreB colored by domain. Domains IA, IB, IIA, and IIB are labeled, and the N and C termini are shown. Nucleotide (ATP) is shown in gray, and the residues involved in membrane binding are indicated.