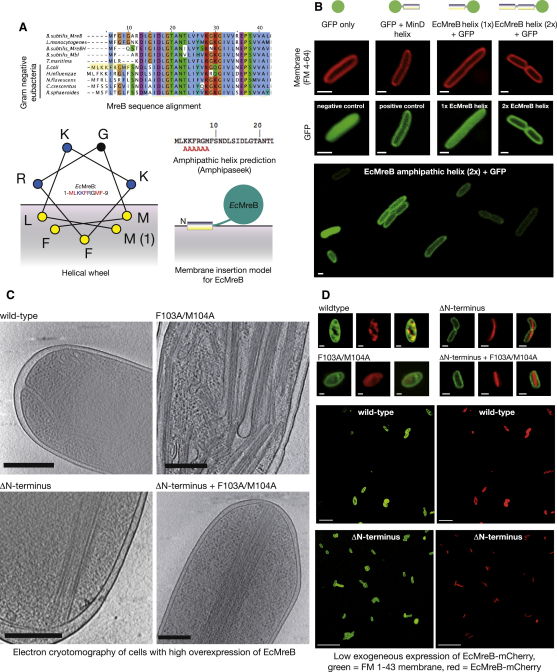
Figure 3.
EcMreB Has a Small N-Terminal Amphipathic Helix that Is Necessary and Sufficient to Confer Membrane-Binding Activity
(A) (Top) Multiple sequence alignment of MreBs. Sequences from Gram-negative organisms are highlighted and carry an additional N-terminal sequence. (Left) Helical wheel showing the view along the first nine residues of EcMreB. Hydrophobic residues are clustered on one side to form a membrane binding surface. Residues are colored by properties: hydrophobic, yellow; basic, blue; uncharged, black. (Right) Results of amphipathic helix prediction on EcMreB from AMPHIPASEEK software (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_amphipaseek.html). Red “A”s indicate predicted amphipathic helical regions. Below is the membrane insertion model for EcMreB showing the N-terminal amphipathic helix.
(B) Confocal microscopy images showing the localization pattern of GFP alone (pRSET/EmGFP) or with an additional amphipathic helix from MinD (pJS110) or one or two copies of the amphipathic helix from EcMreB (pFE356 and pJS111, respectively). Red, FM4-64 membrane stain; green, GFP. Scale bar, 1 μM.
(C) Digital sections through 3D electron cryotomography reconstructions of E. coli cells in which EcMreB (WT or mutant) has been expressed to high levels (WT EcMreB pFE57, double mutant F103A/M104A pJS107, pJS108, ΔN terminus/F103A/M104A PJS109). Protein expression levels are roughly as shown by whole-cell SDS-PAGE analysis in Figure S3C. Scale bar, 250 nm.
(D) Confocal microscopy images showing wild-type E. coli cells containing low-level overexpression of wild-type and mutant EcMreB-mCherrySW. Wild-type EcMreB-mcherrySW pFE363, EcMreB-ΔN F103A/M104A-mcherrySW pFE364, EcMreB-F103A/M104A-mcherrySW pFE365, EcMreB-ΔN mcherrySW pFE366. Green, FM1-43 membrane stain; red, mCherry. Scale bar, 1 μM.