Figure 4.
The Amphiphatic Helix of MreB Is Required for Cell Shape Maintenance in E. coli
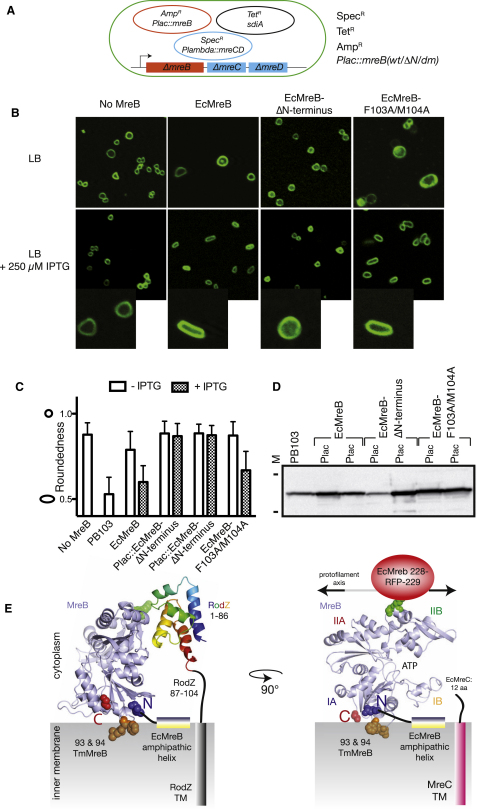
(A) Schematic diagram showing the genetic background. An MreBCD knockout strain (mreBCD < > frt, FB17) carries three plasmids: a plasmid constitutively expressing transcription factor SdiA (pFB112, tetR) that enhances the expression of FtsQAZ, a plasmid that carries mreCD downstream of a lambda promoter and temperature-sensitive repressor (pFB124, specR), and a plasmid expressing the mreB variants under a lac promoter (pFB209, wild-type MreB; pFE379, EcMreB-ΔN terminus; pFE380, EcMreB-F103A/M104A, all ampR).
(B) The N-terminal amphipathic helix of MreB is essential for shape maintenance in E. coli. Confocal microscopy images of strain FB17/pFB112/pFB124 (ΔmreBCD/tet sdiA/ aadA clts plambda::mreCD, first column) and of the same strain transformed with pFB209 (plac::EcmreB, second column), pFE379 (plac::EcMreB-ΔN, third column), and pFE380 (plac::EcMreB-F103A/M104A, fourth column). MreB versions were induced with 250 μM IPTG. Cells that express functional MreB (EcMreB and EcMreB-F103A/M104A) revert to rod shape, whereas expression of EcMreB-ΔN results in misshapen cells. Cells were grown for 6–7 hr in LB at 37°C supplemented with ampicillin and spectinomycin and stained with FM4-64 prior to visualization with a Zeiss confocal laser scanning microscope LSM 510.
(C) Cell shape distribution. The graph presents width/length ratios as a measure of cell roundedness (performed computationally with ImageJ). Perfect round cells have a value of 1.0 and perfect rods a value around 0.6 for E. coli. Strains and growth conditions were the same as in (B), and strain PB103 was used as a wild-type control. The graph is based on three independent experiments, with total number of cells measured as follows: no MreB (FB17/pFB112/pFB124), n = 114; PB103, n = 87; EcMreB (FB17/pFB209/pFB124), n = 76 (−IPTG), n = 82 (+IPTG); Plac::EcMreB-ΔN (FB17/pFE379/pFB124), n = 131 (−IPTG), n = 150 (+IPTG); Ptac::EcMreB-ΔN (FB17/pFE377/pFB124), n = 106 (−IPTG), n = 52 (+IPTG); EcMreB-F103A/M104A (FB17/pFE380/pFB124), n = 98 (−IPTG), n = 143 (+IPTG). Error bars represent standard deviations.
(D) Western blot showing the levels of MreB variants in extracts from the corresponding strains used in Figure 4C. Equal amounts of cells were loaded and MreB was detected using affinity-purified α-MreB antibodies. The positions of the 35.9 and 52.7 kDa standards are indicated.
(E) Schematic diagram showing the position of MreB on the membrane and its interactions with RodZ (known from a cocrystal structure, van den Ent et al., 2010) and the C-terminal peptide of EcMreC. The membrane insertion loop and EcMreB amphipathic helix are shown. The position of mCherrySW used to construct a functional fusion protein in the internal loop is shown, and the close positions of the N and C termini to the membrane reveal why previous N- or C-terminal fusion proteins were likely nonfunctional.