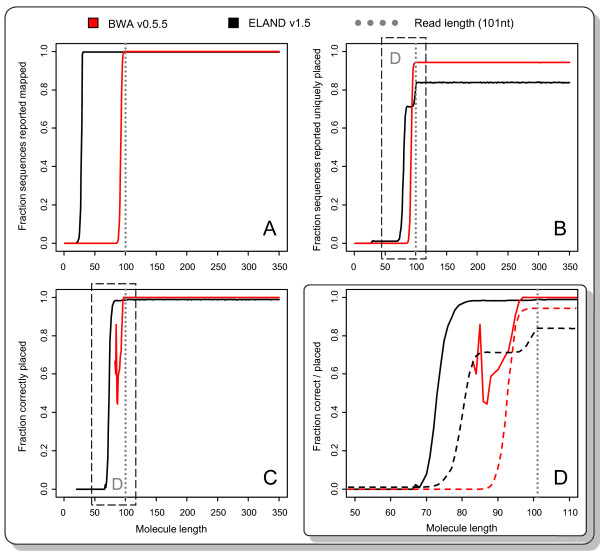
Figure 3.
Effects of adapter sequence inclusion on mapping. Untrimmed adapter sequence at the read ends can interfere with alignment/mapping. We simulated 101-cycle human genomic shotgun reads for an Illumina Paired End library with 10,000 reads for every adapter starting point between 1 to 350nt, and the error profile observed for an actual run of this length. On this data set, we tested how ELAND and BWA are affected by inclusion of adapter sequence: (A) ELAND requires only a fixed seed (here 32nt) in the beginning of the read. Adapters beginning after this seed region may therefore have no effect on the output. ELAND reports 98% successful mappings for all simulated reads of at least 30nt insert size (2nt of adapter sequence being compensated by 2 mismatches allowed in the seed), BWA only reports 98% successful mappings for reads with an insert size of at least 97nt. (B) Frequently only uniquely placed molecules are considered in data analysis. ELAND reports the first uniquely placed fragment for 20nt insert size. BWA reports the first three uniquely placed fragments (mapping quality above 20) for an insert size of 83nt. (C) All uniquely placed reads reported by ELAND up to an insert length of 67nt are placed incorrectly (when comparing to the coordinates the sequence was extracted from), as is one of the 3 reported by BWA for an insert size of 83nt. When requiring 98% correct placements, ELAND handles up to 14nt of adapter (83nt insert size), while BWA can only compensate with mismatches for 4nt of adapter sequence (97nt insert size). (D) For analysis purposes, BWA shows the better performance due to the lower number of false positive placements. Moreover, for an insert size of at least the read length (i.e. no adapters interfering with the alignment), BWA reports 99.999% of uniquely placed reads (94.2% of all reported alignments) at the designated genomic positions, while ELAND only reports 98.757% of the uniquely placed reads (83.8% of all reported alignments) at the correct position.