Figure 6.
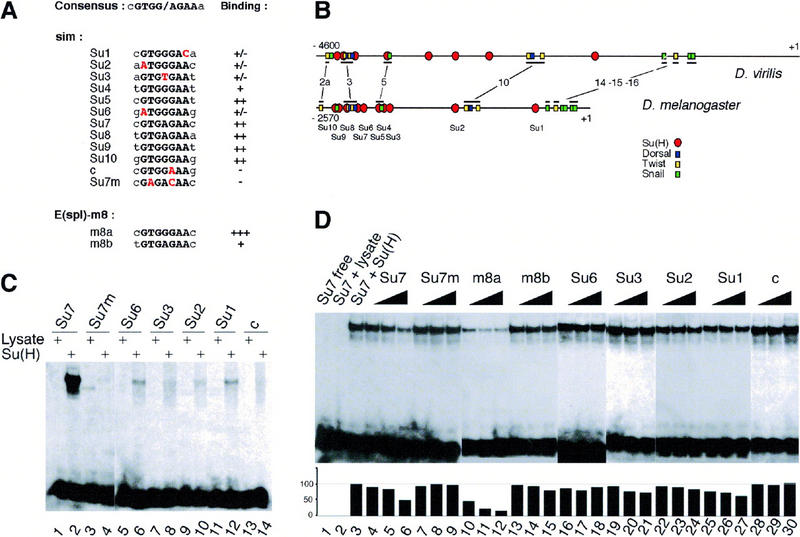
Identification of Su(H)-binding sites in the sim regulatory region. (A) Alignment of the predicted Su(H)-binding sites (Su1 to Su10) contained within a 2.8-kb upstream regulatory region of sim to the consensus Su(H)-binding site. The core consensus is shown in black uppercase letters; the two residues flanking the core consensus are less conserved. Sites Su4, Su5, Su7, Su8, Su9, and Su10 perfectly match the core consensus. Sites Su1, Su2, Su3, and Su6 differ at one conserved position (as indicated in red). Putative site c differs at a position shown previously to be essential for CBF-1/RBP-Jk binding (Tun et al. 1994). The sim regulatory sequence contains no other sites differing by less than two conserved nucleotides. Two binding sites from the Enhancer of split m8 gene [E(spl)-m8] were used as positive controls (Bailey and Posakony 1995; Lecourtois and Schweisguth 1995). For each putative site, the relative binding affinity, as estimated from gel shift assays, is indicated on the right. (+++, ++, +, +/−) Very high, high, medium, and weak binding affinity, respectively; (−) no detectable binding. Site c, which differs from the consensus at a strictly conserved position, did not bind Su(H). (B) Schematic diagrams of the upstream regions of the sim genes from D. virilis and D. melanogaster (Kasai et al. 1998). The position of the predicted Su(H)-binding sites is shown relative to the predicted Snail-, Twist-, and Dorsal-binding sites. The conserved regions that include known binding sites are underlined. These correspond to regions 2a, 3, 5, 10, 14, 15, and 16 described in Kasai et al. (1998). Four binding sites predicted to bind strongly Su(H) (Su9, Su8, Su7, and Su5) appeared to be clustered with predicted Snail-, Twist-, and Dorsal-binding sites in both D. virilis and D. melanogaster. Nucleotide numbering refers to the translation initiation codon. (C) Gel retardation analysis of Su(H) binding to putative sites from the sim regulatory region. Radiolabeled 17-mer oligonucleotides centered around putative Su(H) binding sites were tested for their ability to form retarded complex with Su(H) in an EMSA. One site perfectly matching the core consensus, Su7, as well as all the sites differing by one nucleotide to the core consensus (Su6, Su3, Su2, Su1, and c) were analyzed. For each probe, free lysate was used as a negative control (lanes 1,3,5,7,9,11,13). In vitro translated Su(H) proteins bound strongly to Su7 (lane 2). Weak binding was also observed with Su6, Su3, Su2, and Su1 (lanes 6,8,10,12, respectively). No detectable binding was observed with putative site c (lane 14). Binding specificity was demonstrated with an oligonucleotide containing two mutations in the Su7 site, Su7m (lane 4). These results are consistent with the binding specificity displayed by the mouse homolog of Su(H) (Tun et al. 1994), as the Su7m and c sites are the only ones that contain nucleotides differing from the consensus at strictly conserved position. (D) Determination of relative binding affinities by competition EMSA. Increasing amounts (5×, 10×, and 20×) of nonlabeled oligonucleotides were tested for their ability to compete with the formation of radiolabeled Su7–Su(H) complex (lanes 1–3). The m8a (lanes 10–12) and Su7 (lanes 4–6) oligonucleotides efficiently competed the binding of Su(H) to Su7. The m8b (lanes 13–15), Su6 (lanes 16–18), Su3 (lanes 19–21), Su2 (lanes 22–24), and Su1 (lanes 25–27) oligonucleotides competed only weakly. The Su7m (lanes 7–9) and c (lanes 28–30) oligonucleotides did not show significant competition activity. The plot underneath the EMSA gel shows the quantitation of the radioactivity contained within retarded complexes as measured by PhosphorImager analysis. The radioactivity measured in the absence of specific competitor was chosen as the 100% reference (lane 3).
