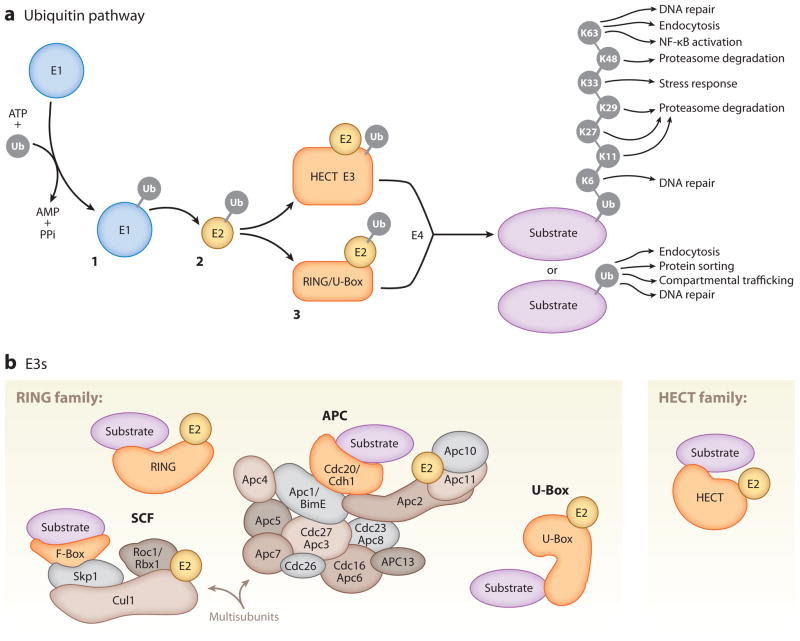
Figure 2.
Components of the ubiquitin (Ub) pathway. (a) Substrate ubiquitination requires a three-step enzymatic reaction. In step 1, a ubiquitin-activating enzyme (E1) activates free ubiquitin, a process in which the C-terminal glycine residue of ubiquitin is coupled to a cysteine residue of the E1 via a thioester linkage. In step 2, the activated ubiquitin is transferred to a cysteine residue of a ubiquitin-conjugating enzyme (E2) and linked via a thioester linkage. In step 3, a ubiquitin ligase (E3) transfers ubiquitin to the substrate via an amide isopeptide linkage to the ε-amino group of the substrate lysine (Hershko & Ciechanover 1998). In some cases, a polyubiquitin ligase (E4) coordinates with a ubiquitin E3 to lengthen ubiquitin chains (Hoppe 2005). Ubiquitin can be attached to lysine residues of previously conjugated ubiquitins in multiple polyubiquitin chain configurations (K6, K11, K27, K29, K33, K48, K63) to regulate various functions. (b) Ubiquitin E3s are divided into the RING (really interesting new gene) and HECT (homologous to E6-associated protein C terminus) domain families, among others. The RING domain family is the largest family of ubiquitin E3s. RING ubiquitin E3s function as single subunits or in a multisubunit complex. Illustrated here are the best-studied multisubunit RING ubiquitin E3s, the Skp1/Cullin/F-box (SCF) and anaphase-promoting complex/cyclosome (APC/C) complexes. U-Box proteins are a distinct set of RING-like ubiquitin E3s.