Abstract
Thyroid hormone action can be customiZed on a cell-specific fashion through the controlled action of the deiodinase group of enzymes, which are homodimeric thioredoxin fold containing selenoproteins. Whereas the type II deiodinase (D2) initiates thyroid hormone signaling by activating the pro-hormone thyroxine (T4) to the biologically active T3 molecule, the type III deiodinase (D3) terminates thyroid hormone action by catalyzing the inactivation of both T4 and T3 molecules. Deiodinases play a role in thyroid hormone homeostasis, development, growth and metabolic control by affecting the intracellular levels of T3 and thus gene expression on a cell-specific basis. Whereas both Dio2 and Dio3 are transcriptionally regulated, ubiquitination of D2 is a switch mechanism that controls D2 activity and intracellular T3 production. The hedgehog-inducible WSB-1 and the yeast Doa10 mammalian ortholog TEB4 are two E3 ligases that inactivate D2 via ubiquitination. Inactivation involves disruption of the D2:D2 dimer and can be reversed via two ubiquitin-specific proteases, USP20 and USP33, rescuing catalytic activity and T3 production. The ubiquitin-based switch mechanism that controls D2 activity illustrates how different cell types fine-tune thyroid hormone signaling, making D2 a suitable target for pharmacological intervention. This article reviews the cellular and molecular aspects of D2 regulation and the current models of D2-mediated thyroid hormone signaling.
Introduction
Thyroid hormones play critical developmental and metabolic roles in all vertebrates (Cheng et al., 2010). Thyroxine (T4) is the most abundant product of the thyroid gland, which is a minimally active form of thyroid hormone that must be converted to T3 (3,5,3’-triiodothyronine) in order to gain full biological activity. This activation pathway is mediated by a group of enzymes known as iodothyronine deiodinases (D1 and D2), which also includes an inactivating deiodinase, the type 3 deiodinase (D3), that inactivates both T4 and T3 (Bianco et al., 2002). These deiodinase-mediated pathways are intracellular and thus depend on the intracellular availability of the different substrates (Friesema et al., 2006). Access to the intracellular compartment is mediated by four different families of transport proteins have been shown to be involved in the traffic of iodothyronines across the plasma membrane, with different pattern of tissue expression (Visser et al., 2011). The monocarboxylate 8 (MCT8) is probably the most relevant transporter as mutations in the MCT8 protein have been associated with variable levels of mental retardation in humans combined with lack of speech development, muscle hypotonia and endocrine dysfunctions (Visser et al., 2011, Dumitrescu et al., 2004, Friesema et al., 2006). Intracellular T3 binds to two different nuclear thyroid hormone receptors (TRs), TRα and β, that mediate the biological effects of thyroid hormone via transcriptional control of multiple sets of T3-responsive genes (Cheng et al., 2010). The modern paradigm of thyroid hormone action recogniZes that D2 and D3 play antagonistic roles in a number of settings, with D2 increasing thyroid hormone signaling and D3, silencing it, relatively independently of plasma T4 or T3. Thus, at any given moment, thyroid hormone signaling in different tissues and cells can be amplified or dampened, according to the local expression of deiodinases (Gereben et al., 2008).
The role played by the deiodinases is physiologically relevant, playing a role in various aspects of mammalian physiology such as the maintenance of plasma T3 concentration (Bianco et al., 2002), TSH and TRH feedback regulation (Larsen, 1982, Christoffolete et al., 2006, Fekete et al., 2004) and in the clearance of sulfated iodothyronines (Schneider et al., 2006). In addition to homeostatic processes, deiodinases also play a role during development (Galton, 2005) and in diseases states (Huang and Bianco, 2008). Many of these mechanisms have been validated using deiodinase null mice (St Germain et al., 2005, Hernandez et al., 2006).
The cloning of all three deiodinases identified the presence of the rare amino acid selenocysteine (Sec) located in the catalytic site (St Germain and Galton, 1997). Like with the other selenoproteins, deiodinases undergo a unique synthetic mechanism that involves reprogramming of the stop codon, UGA, to a Sec amino acid (Berry et al., 1991). This takes place with a coordinated effort between the ribosomal machinery and a evolutionary conserved stem loop-shaped RNA element that is found at the 3’UTR of all mammalian selenocysteine proteins (Berry et al., 1991). This element is called the selenocysteine insertion sequence (SECIS) and functions as a binding site of several accessory proteins that recruit a Sec-charged tRNA and mediates the anti-termination at the UGA codon. This allows the incorporation of a Sec molecule into the selenoprotein growing polypeptide chain.
Subsequent sequence analysis coupled with hydrophobic cluster analysis (HCA), show that the deiodinases share an overall 50 % sequence similarity. The majority of this similarity lies within the conserved thi-oredoxin-fold (TRX) domain with the classical four-stranded anti parallel β-sheets and two alpha helixes, composed of βαβ and ββα motifs (Callebaut et al., 2003). Similar to other TRX-fold containing proteins, secondary structures may lie within these βαβ/ββα motifs, where the deiodinases contain a unique conserved secondary sequence that shares a high degree of similarity (47 % for D1 and D3; 60 % for D2) with the lysosomal α-L-iduronidase (IDUA) that processes the sulfated form of the α-L-iduronic acid, which is strikingly similar to T4 and T3 (Callebaut et al., 2003).
All three deiodinases are homodimers and the data indicate that dimerization is required for full catalytic activity (Leonard et al., 2001, Curcio-Morelli et al., 2003a, Sagar et al., 2008, Sagar et al., 2007). This has been confirmed by co-immunoprecipitation of differently tagged D1:D1, D2:D2 or D3:D3 homodimers. Dimerization involves the TM domain and also structures in the globular domains of the deiodinases, and may include disulfide bonds as well (Curcio-Morelli et al., 2003a). Remarkably, there is a small level of heterodimerization between D3:D1 or D3:D2, the significance of which remains unknown (Sagar et al., 2008, Curcio-Morelli et al., 2003a).
D1 is located in the plasma membrane and has a slow turnover rate of about 8 h (Baqui et al., 2003). D1 is anchored in the plasma membrane via a single transmembrane domain (TM) with its catalytic globular domain facing the cytosol (Sagar et al., 2008). The Diol gene is highly sensitive to T3, with a ~ 175-fold induction during the transition from hypothyroid to hyperthyroid states, thus functioning as an indicator of systemic thyroid status (Zavacki et al., 2005). Although capable of T4 to T3 deiodination, studies with D1 knock out (KO) mice indicate that D1 plays a scavenger role, preferentially deiodinating sulfated forms of iodothyronines in the process of being eliminated in the bile and urine (Schneider et al., 2006).
D2 is considered the main T4-activating enzyme, given its high substrate affinity (Km ~ 2 nM T4 vs. Km ~ 1 µM T4 for D1). D2-mediated T3 production happens intracellularly. Subsequently, T3 exists the cells and enters the plasma compartment, being responsible for 70 % of all extrathyroidal T3 production in healthy humans (Bianco et al., 2002). D2 is a classical type-1 membrane protein residing on the endoplasmic reticulum (ER) membrane, with a half-life of ~ 45 minutes (Baqui et al., 2003). Its relatively short half-life is due to ubiquitination and proteasome uptake, a feature that is accelerated by D2 interaction with its natural substrate, T4 (Gereben et al., 2000). This constitutes a powerful homeostatic mechanism that minimiZes changes in the levels of T3, the active form of thyroid hormone, in the face of fluctuating T4 levels, such as during iodine deficiency, for example. D2’s catalytic domain faces the cytosol and its TM domain is embedded into the ER membrane with the NH2-termini in the ER lumen (Baqui et al., 2000). Residency in the ER is fundamentally critical for D2, as it is clear that D2-generated T3 is transferred to the TR-containing nuclear compartment. In D2 expressing cells, most TR-bound nuclear T3 is made intracellularly via the D2 pathway. D2 expression is regulated on time- and cell-specific fashion by a combination of transcriptional modulation of the Dio2 gene, post-transcriptional mechanisms regulating Dio2 mRNA stability, and by post-translational mechanisms such as ubiquitination (Bianco and Kim, 2006).
D3 is highly expressed in the placenta where it plays a critical role in protecting the developing embryo from excessive TH levels (Dentice and Salvatore, 2011, Gereben et al., 2008). D3 is an obligatory innerring deiodinase with a nanomolar affinity for both T4 (Km ~ 30 nM) and T3 (Km ~ 6 nM) and a half-life of about 12 h (Baqui et al., 2003, Salvatore et al., 1995). The mature form of D3 resides in the plasma membrane where it is rapidly internaliZed and recycled between the plasma membrane and early endosomes. Although initial characterization of D3 topology put the catalytic active site in the extracellular space (Baqui et al., 2003), functional studies indicate that substrates must have access to the intracellular compartment in order to be metaboliZed by D3 (Friesema et al., 2006).
Thyroid hormone signaling and D2
Thyroid hormone action and tissue development
During vertebrate development, thyroid hormone action occurs in a time and cell dependent manner (Galton, 2005, Galton, 1992). This is achieved through the coordinated actions of D2 and D3. In general, D3 predominates in most tissues during early development, preventing untimely exposure of the rapidly developing cells to thyroid hormone. As the embryo matures, D3 expression subsides and D2 rises substantially, increasing thyroid hormone signaling until after birth and through adulthood.
Sensory organs development
All components of the thyroid hormone signaling pathway are expressed during the chicken and mouse retina development as well as being a determinant factor for proper cochlear development (Ng et al., 2010, Trimarchi et al., 2008, Ng et al., 2004). Although Dio2−/− mice present euthyroid circulating levels of T3, they present a cochlear phenotype similar to TRβ-deficient or hypothyroid mice, developing auditory and morphology defects consistent with lack of thyroid hormone action. In fact, prevention of the defective cochlear development in Dio2−/− mice was achieved by post-natal supplementation with a high dose of T3 (Ng et al., 2004). Similarly, the development of the mouse retina, which takes place during the neonatal period, is also highly dependent on timed thyroid hormone action. Hypothyroid mice fail to develop proper expression of different opsins (M and S) while the number of cones is normal, a phenotype that is reversed/corrected by treatment of neonates with exogenous doses of T3 (Lu et al., 2009).
Skeletal development
Thyroid hormone actions have been linked to proper skeletal growth and function. Initially, it was observed that D2 was expressed in chondrocyte and osteoblast cell lines during the differentiation process, where thyroid hormone was required for proper cellular maturation (Gouveia et al., 2005, Miura et al., 2002). Furthermore, in the chicken bone growth plate, the identification of a sonic hedgehog driven mechanism regulating the levels of D2 and D2-mediated T3 production via D2 ubiquitination revealed the key role that proper local and timed thyroid hormone action plays in bone development (Dentice et al., 2005). Interestingly, patients carrying the Thr92Ala Dio2 polymorphism, which has been shown to have lower D2 activity (Canani et al., 2005), have decreased bone mineral density and increased bone turnover (Heemstra et al., 2010). Studies with the Dio2−/− mouse indicate that its bones are more susceptible to fracture accompanied by a 2-fold reduction in bone formation (Bassett et al., 2010). Besides D2, the thyroid hormone inactivating enzyme, D3, is also expressed in developing osteocytes. D3 expression is observed in the fetal skeleton, where it generates a pro-proliferative environment by creating an intracellular hypothyroid state that is required for proper bone development (Capelo et al., 2008).
Regulation of the hypothalamus-pituitary-thyroid axis
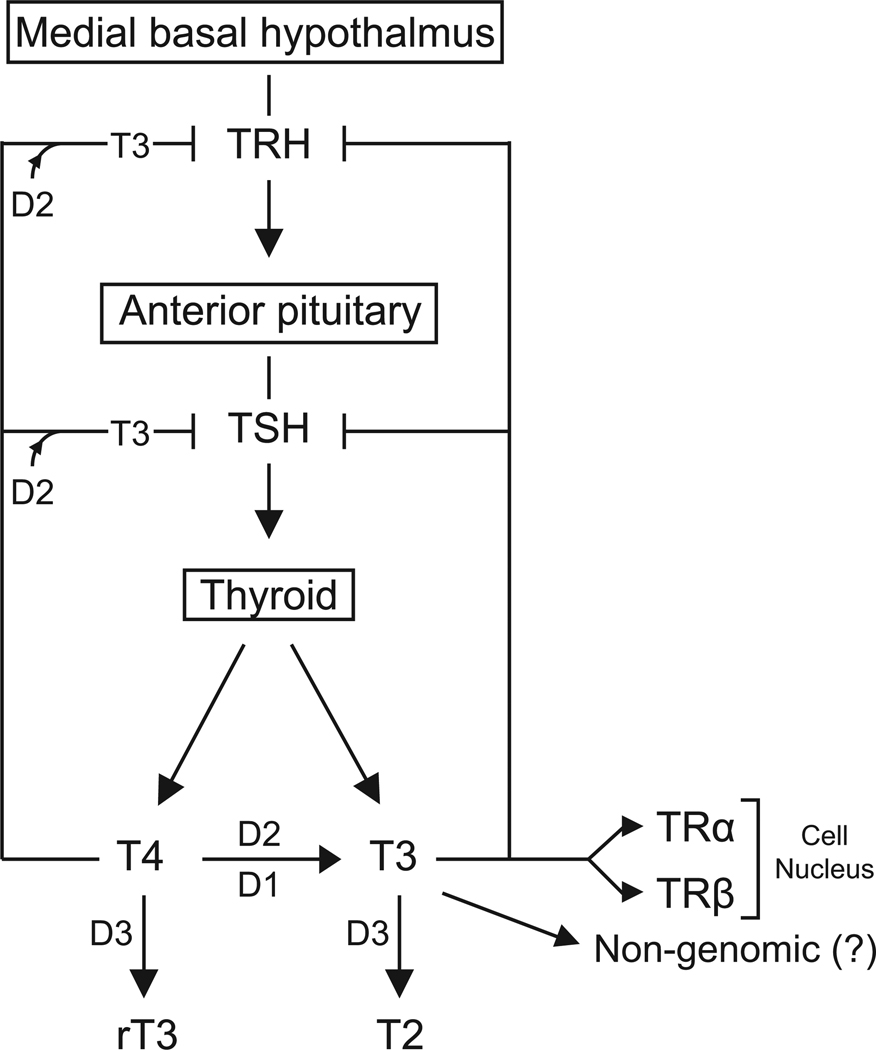
The thyroid-stimulating hormone (TSH) and the thyrotropin-releasing hormone (TRH) promote thyroid gland activity and thyroid hormone production. TRH is produced by paraventricular nucleus neurons, is released in the portal blood and positively regulates pituitary TSH secretion. An elevation in TSH secretion increases thyroid hormone synthesis and secretion that elevates the circulating levels of thyroid hormone, closing a negative feedback loop that inhibits both TRH and TSH secretion (Chiamolera and Wondisford, 2009). Given that T4 is only minimally active, D2 plays a central role in this regulatory loop, as it transduces the elevation in plasma T4 into intra-pituitary levels of T3 for proper regulation of TSH expression (Larsen, 1982, Christoffolete et al., 2006). An increase in circulating T4 and the consequent increase in intracellular T3 concentration in the pituitary thyrotrophs (cells that produce TSH), shuts down TSHb gene expression, while a drop in circulating T4 results in an opposite effect in TSHb gene expression (Christoffolete et al., 2006). Supporting the regulatory role D2 plays on the HPT axis, inactivation of the Dio2 gene in mice leads to central resistance to T4 (Schneider et al., 2001), a phenotype that is reproduced in mice treated with amiodarone, a D2 inhibitor commonly used as an antiarrhythmic drug (Rosene et al., 2010). A similar role for D2 in TRH regulation has been proposed (Fekete et al., 2004). In this case, D2 is expressed in the tanycytes, which are specialiZed ependimal cells located in the walls of the III ventricle. It is conceivable that D2-generated T3 in tanycytes mediate a negative feedback at the level of the paraventricular TRH neurons via paracrine mechanisms. In fact, recent studies have shown that this type of hormonal signaling involving deiodinase-pathways is possible (Freitas et al., 2010). A role for deiodinases in the regulation of the hypothalamus-pituitary-thyroid axis in humans, including D2, is supported by the observation that thyroid hormone homeostasis and TSH feedback mechanism is disrupted in patients with SECIS-BP2 mutations, a key protein involved in the selenoprotein synthesis (Dumitrescu et al., 2005).
Adaptative thermogenesis and metabolic control
When enduring cold temperatures, small mammals trigger an integrated metabolic response in order to generate heat via uncoupling mitochondrial respiration, a process known as non-shivering thermogenesis or adaptative thermogenesis (Silva, 2011). The release of norepinephrine (NE) in the brown adipose tissue (BAT)/sympathetic nervous system (SNS) synaptic terminals increases the intracellular production of cAMP and up-regulation of cAMP responsive genes, including the metabolic drivers peroxisome proliferator-activated gamma co-activator 1 alpha (PGC-1α) and D2. The increase in D2 activity (~ 26-fold) and of local D2-generated T3, is critical for proper BAT functions, as it increases the responsiveness of the BAT to NE signaling and directly mediates the increase in total levels of the uncoupling protein 1 (UCP-1) (Carvalho et al., 1991, Bianco et al., 1992). In addition to Dio2 transcriptional up regulation, D2 activity levels are kept at high levels due to a cAMP-mediated rise in the expression of the ubiquitin specific protease 33 (USP-33 or VDU-1), which prolongs D2 half-life by deubiquitination of D2 (Curcio-Morelli et al., 2003b).
The role that D2 plays in BAT is underscored by the fact that animals lacking D2 expression (Dio2−/−) are cold intolerant and develop hypothermia after 12 h of cold exposure due to deficient BAT function (de Jesus et al., 2001). In addition, Dio2−/− mice kept on a high fat diet develop obesity and severe hepatic steatosis when at thermoneutrality, illustrating the critical role played by D2 in metabolic control (Castillo et al., 2011). The specific mechanism underlying these phenotypes has its origins during BAT development in the Dio2−/− mouse, in which there is low expression of UCP-1 and PGC1α (Hall et al., 2010). In addition, Dio2−/− primary brown adipocytes have an impaired differentiation capacity and a decreased cellular machinery to cope with oxidative stress, leading to an increase in levels of oxidative stress/ROS (Hall et al., 2010). Interestingly, Dio2−/− primary brown adipocytes also show impaired Akt phosphorylation, a phenotype that is reversed by treatment with ascorbic acid, a potent anti oxidant. This increase in ROS levels is connected with D2 inactivation, as chemical inactivation of D2 via addition of rT3 – a condition that rapidly decreases D2 protein levels - leads to similar decrease in Akt phosphorylation as seen in the Dio2−/− cells (Hall et al., 2010). A developmental role for D2 was also suggested in the skeletal muscle, as D2 expression is found in both humans and murine muscle tissue (Grozovsky et al., 2009) and in cultured muscle myoblasts, D2 and D2-generated T3 were shown to be necessary for proper myogenic development (i.e. Myo D induction) and regeneration, while this regulation was dependent interaction of the transcription factor Forkhead box O3 (FoxO3) with the Dio2 gene promoter region (Dentice et al., 2010).
Molecules regulating the D2 pathway
Molecules belonging to the family of flavonols, a small class of polyphenolic compounds widely found in today’s diet, such as resveratrol, quercetin, fisetin and kaempferol (KPF), have received a great deal of attention due to their proposed anti-cancer properties and also control mammalian metabolism by activation of the sirtuin pathway (Baur and Sinclair, 2006). A screening for xenobiotic compounds that induce D2 expression and intracellular D2-generated T3 identified KPF as a potent activator of the D2 pathway (da-Silva et al., 2007). KPF treatment increased D2 activity up to 50-fold in various cell models via transcriptional mechanisms, while increasing oxygen consumption by 30 % and the expression of key metabolic genes such as PGClα, CPT-1, uncoupling protein 3 (UCP-3), mitochondrial transcription factor 1 (mTFA), including also the Dio2 gene (da-Silva et al., 2007).
Bile acids along with cholesterol and other lipids compose the bile, which is secreted by hepatocytes and stored in the gall bladder. Bile acids have been promoted from molecules that assisted on the digestion of fatty nutrients to major integrators of metabolic signals and are now linked with activation of the mitogen-activated kinase pathways, FXRα nuclear receptors and the G-coupled protein receptor TGR5 (Houten et al., 2006, Watanabe et al., 2006). The metabolic role of bile acids became clear when animals fed a high fat diet supplemented with cholic acid, gained as much weight and fat as animals kept on a chow diet. Dietary cholic acid supplementation also reversed the body weight gain in animals kept on a high-fat diet for 120 days. All these effects were mediated via TGR5 activation independently of FXRα activation given that treatment with the FXR agonist GW4064 did not decrease weight gain (Watanabe et al., 2006). Dietary bile acid supplementation also increased oxygen consumption that coincided with an increase in BAT expression of PGC1α and β, CPT-1, UCP-3 and D2 (Watanabe et al., 2006). In fact, the protective metabolic effects of bile acid diet supplementation were found to be D2-dependent, given that they were lost in Dio2−/− mice. Furthermore, different bile acid molecules that act as TGR5 ligands were capable of stimulating Dio2 promoter and enzymatic activity several fold (Watanabe et al., 2006). The fact that in murine BAT and also in human skeletal muscle there is co-expression of TGR5 and D2, points out to the significant role that the bile acid-TGR5-D2 pathway plays in metabolically relevant tissues, being a strong candidate for pharmacological intervention for treatment of metabolic syndrome (Watanabe et al., 2006, Pols etal., 2010).
D2 is an ER-resident protein
D2 is retained in the ER
The ER is a dynamic cellular organelle that among various functions is involved in protein synthesis and degradation (Schröder, 2008). Proteins targeted to the ER membrane have a specific targeting signal, best characteriZed by the presence of di-lysine motifs within their amino acid sequence, where mutation of such motifs leads to a change in protein cellular localization (Hardt and Bause, 2002). The D2 amino acid sequence contains 15 lysine residues but combinatorial Lys to Arg mutation was not able to change the D2’s ER residency. Only when the transmembrane domain of D2 was deleted, D2 was found to be a cytosolic protein, which suggests that the ER retention signal lies within the transmembrane domain (Zeöld et al., 2006). During the protein maturation process in the ER lumen, ER resident proteins can be transported to the Golgi cysternae for post-translational modifications and then are transported back to the ER (Spang, 2009, Schröder, 2008). To determine whether D2 would cycle from the ER to the Golgi system, ER- or Golgi-specific glycosylation signals were fused to either termini of the D2 molecule. While fusion of the Golgi-specific signal (YTPPP) did not yield any evidence of glycosylation, fusion of the ER-specific signal (NKT) onto the amino termini of D2 showed evidence of protein glycosylation (Zeöld et al., 2006). This observation indicated that D2 did not reach the Golgi cysternae system, being actively retained at the ER.
When proteins fail to fold properly or become misfolded within the ER lumen, they are targeted for degradation by a mechanism called endoplasmic reticulum associated degradation (ERAD) (Vembar and Brodsky, 2008, Schröder, 2008). Due to that D2 resides in the ER, one could hypothesiZe that D2 protein instability and short half-life is due to its sub-cellular localization and the physical proximity to the ERAD machinery. To test this hypothesis, a D2 protein lacking its transmembrane domain was fused to the carboxyl termini of the long-lived (half-life of ~ 5 days) plasma membrane resident protein Na+/I− symporter (NIS) (Dohán et al., 2003). This followed the rationale that by fusing the globular domain of D2 to a plasma membrane protein would distance the ERAD machinery from D2, thus making it possible to analyZe intrinsic characteristics of the D2 protein. As expected, the D2-NIS chimeric protein segregated to the plasma membrane and displayed a much shorter half-life (~ 4 h) when compared to the wild type NIS, whereas deletion of D2’s 18-aa instability domain prolonged D2/NIS protein half-life (Zeöld et al., 2006). Similar results were observed when D2 was fused to the stable ER resident protein Sec62. Taken together, these results show that, independently of cellular location, D2 contains intrinsic motifs that render instability and a short half-life to the D2 protein.
Dimerization - it takes two to deiodinate
An analysis of the amino acid sequence of deiodinases predicts a molecular weight ranging between 29–33 kDa, but in gel filtration studies identified the presence of deiodinases in higher molecular weight complexes ranging from 44–200 kDa (Safran and Leonard, 1991). This suggests that deiodinases could be part of multi-protein complexes or were organiZed in multimeric forms. Accordingly, when a mutant inactive rat D1 isoform was co-expressed in endogenously D1 expressing porcine LLC-PK1 cells, D1 activity was lost when rat D1 was immunoprecipitated, suggestive of a dimeric form of active D1 (Leonard et al., 2001).
Using an approach that couples protein dimerization with fluorescence resonance energy transfer (FRET), Sagar et al sought to characteriZe the D2-D2 dimer formation and at the same time study the dynamics of the substrate-induced D2 degradation (Sagar et al., 2007). CFP and YFP labeling of the carboxyl or the amino termini of the D2 molecule made it possible to study D2 dimerization at the globular and TM domains (Sagar et al., 2007). Removal of the TM domain in D2 generates an inactive truncated ΔD2. When co-expressed with wild type D2, there is ΔD2:D2 dimerization as detected by FRET along with minimal levels of enzyme activity (Sagar et al., 2007). This observation indicates that the globular domain alone is sufficient for enzyme dimerization and function, as long as one of the counterparts is anchored to the ER membrane. The TM contains potential dimerization motifs, as sequence analysis shows that the D2 transmembrane domain is helix-shaped and due to the presence of positively charged amino acids it would be stable when inserted onto the ER membrane, thus supporting an α-α dimeric structure (Sagar et al., 2007). The critical observation that interaction with T4 decreases the FRET signal of carboxyl-tagged D2:D2 dimers, but not the FRET signal of amino-tagged D2-D2 dimers, indicates that interaction with T4 promotes a conformational change in the D2:D2 dimer, which is restricted to the globular domain (Sagar et al., 2007).
D2 and the ubiquitin-proteasome system
The ubiquitin/proteasome system is critical for protein homeostasis
A tight control of protein homeostasis is critical for maintaining the integrity of a number of cellular functions. In addition to the lysosomal pathway, cells also dispose of unwanted proteins after they have been tagged by ubiquitin (Ub) and taken up by the proteasomes (Yang and Klionsky, 2010, Vembar and Brodsky, 2008). Thus, proteins targeted to this system undergo structural modifications that involve conjugation of Ub moieties to Lys residues in the target protein. Ubiquitination is a reversible, cyclic and multi-enzymatic process that can be divided in 3 canonical steps: (i) the initial, ATP-dependent activation of one Ub molecule by an ubiquitin-activating enzyme (E1), (ii) transfer of the activated Ub molecule from the Ub-charged E1 to an Ub conjugating enzyme (E2) and finally, (iii) polyubiquitination of Lys residues within the target protein by an E2 and/or E3 ligase-containing complex (Vembar and Brodsky, 2008). Studies have also identified a new class of enzymes involved in the ubiquitin/proteasome pathway termed E4, which are able to elongate the polyu-biquitin chain of specific substrates (Liu et al., 2010). Finally, with participation of accessory proteins such as HSP90/40 chaperones and the p97 ATPase, the polyubiquinated substrate is lineariZed and shuttled to the proteasome system where it is degraded into oligopeptides (Vembar and Brodsky, 2008).
D2 is ubiquitinated and targeted for the proteasome
The observation that D2 half-life could be prolonged by depletion of rat pituitary cellular ATP levels independently of changes in mRNA levels led investigators to search for the involvement of ATP-dependent degradation pathways (Leonard et al., 1990). The involvement of the proteasome system was confirmed when treatment of D2-expressing cells with the proteasome blocker MG-132 prevented the loss of D2 and prolonged enzymatic half-life by over 50 % (Steinsapir et al., 1998). In addition, exposure of D2 to its natural substrate T4, which accelerates disposal of D2, also was blocked by MG-132. In follow up studies, transiently expressed human D2 was also found to be stabiliZed by treatment with MG-132, thus confirming the post-transcriptional nature of D2’s degradation mechanism (Steinsapir et al., 2000). Furthermore, it was confirmed that substrate interaction with the enzyme’s catalytic site was required for triggering D2 ubiquitination, as a single See l33Ala mutation renders D2 catalytic inactive and resistant to substrate induced ubiquitination (Gereben et al., 2000). The involvement of the ubiquitin system in D2 degradation was demonstrated in TS20 cells that express a temperature sensitive E1 enzyme. This mechanism was found to be D2 specific as the other thyroid hormone activating enzyme, D1, present a much longer half-life (> 12 h) and did not show any evidence of protein ubiquitination (Gereben et al., 2000).
The D2 protein contains 15 Lys residues that could accept conjugation of an ubiquitin molecule. To identify the exact residue of the D2:D2 dimer that is ubiquitinated, a number of combinatorial Lys to Arg mutations were made. Single mutation of residues K237 or K244 led to no changes in D2-ubiquitination rate, whereas mutation of both K237 and K244 resulted in loss of D2 ubiquitination and several fold prolongation of D2 half-life (Sagar et al., 2007). Strikingly, the D2-D2 dimer with the K237R/K244R mutation was resistant to T4-induced conformational change, as exposure of this mutant to increasing amounts of T4 did not lead to changes of FRET signal of the D2:D2 dimer (Sagar et al., 2007).
E2 Conjugases
The identification of mammalian orthologues of two yeast E2 conjugases, UBC6 and UBC7 (Tiwari and Weissman, 2001), made possible for more detailed studies on the proteins involved in the degradation of D2 via the ERAD pathway. Both UBCs are found close or in association with the ER; where UBC6 is physically anchored to the ER membrane via a transmembrane domain and UBC7 is activated and held close to the ER via interaction with the ER membrane protein Cuelp (Bazirgan and Hampton, 2008). More recently, the crystal structure of yeast and mammalian UBC7 has been solved (Arai et al., 2006, Briggman et al., 2005, Cook et al., 1997), while it also has been shown to functionally interact with the E3 ligases Parkin (Imai et al., 2001), HRD1 (Kikkert et al., 2004) and TEB4 (Hassink et al., 2005) on ERAD pathways. Both UBC6 and or UBC7 interact with D2 (Kim et al., 2003, Botero et al., 2002). UBC6 interaction was only seen when it was co-expressed with UBC7, which supports the idea that both conjugases may overlap or co-exist in the D2 degradation complex. In the same study, significant D2/UBC7 interaction was limited only to the carboxyl domain of the D2 protein. In fact, D2 activity is stabiliZed when a truncated carboxyl terminal D2 is co-expressed with wild type human D2, thus confirming previous results that pin pointed the carboxyl termini as the critical instability domain on the D2 protein (Kim et al., 2003). Expression of inactive isoforms of either UBC6 or UBC7 alone had no effect on D2 activity, suggesting that UBC6/7 have overlapping functions. Accordingly, when both inactive isoforms of UBC6/7 were co-expressed, D2 enzymatic activity increased by 2-fold while also preventing the substrate-induced loss in D2 activity and protein levels (Kim et al., 2003).
E3 Ligases
Substrate specificity of the ubiquitination process is ensured by the interaction of the target protein with the E3 ligase family of enzymes. The human genome encodes over 600 different E3 ligases, which are responsible for the ubiquitination of a myriad of substrates from prokaryotes to eukaryotes. Most of such enzymes reside on the ER membrane and are capable of mediating the ubiquitination of either cytosolic or membrane bound substrates (Vembar and Brodsky, 2008). To date, two E3 ligases have been shown to mediate D2 ubiquitination; the sonic-hedgehog (Shh) inducible WD-40 repeat and SOCS box-containing 1 (WSB-1 or SWiP) and the ERAD enzyme TEB4 (the human ortholog of yeast Doa10).
WSB-1, also known as SWiP-1, was initially described in the limb buds and in the somatic mesoderm of the developing chicken embryo. In mice, the WSB-1 gene is located on chromosome 11 at the Wsb1/Nf1 region that interacts with the region Igf2/H19 on chromosome 7 (Ling et al., 2006). WSB-1 expression was characteriZed as one of the earliest markers of limb development along with BMP2, where ectopic sonic hedgehog (Shh) treatment increased WSB-1 expression significantly (Vasiliauskas et al., 1999). WSB-1 has also been linked to pancreatic cancer progression (Archange et al., 2008), neuroblastoma survival (Chen et al., 2006), regulation of the homeodomain-interacting protein kinase 2 (HIPK2) (Choi et al., 2008), as well as being negatively regulated by the sirtuin-1 (SIRT-1) activator resveratrol in human aortic and pulmonary cell models (Hsieh et al., 2010). Initial observations from experiments where over-expression of the WSB-1 resulted on a 2-fold decrease in D2 activity indicated that WSB-1 was a negative regulator of D2. Accordingly, WSB-1 was shown to be an E3 ligase enzyme with capacity to mediate ubiquitination of a D2 molecule (Dentice et al., 2005). Furthermore, detailed structure analysis showed that WSB-1 is part of a multi-protein catalytic complex, termed ECSWSB1, of which are also part of Elongin B and C, Cullin5, RBX1 and the E2 ligase UBC7 (Dentice et al., 2005). The D2-WSB-1 interaction requires an 18 amino acid long domain on the D2 molecule termed “instability loop” and the WD-40 propeller-shaped domain of WSB-1, more specifically the 3rd and 8th propeller-like domains, while the SOCS box mediates interaction with the other members of the ECSWSB1. WSB-1-D2 interplay shed light on the molecular mechanisms involved in the regulation of the developing chicken bone growth plate. In this region, cells perichondrial/periosteal sheaths express both D2 and WSB-1 (Dentice et al., 2005). In this specific setting, expression of WSB-1 is induced by Indian hedgehog (Ihh) secreted from chondrocytes leaving the proliferative pool. This in turn accelerates D2 ubiquitination, decreases local T3 concentration while also transiently decreasing thyroid hormone signaling. This allows the induction of PTHrP expression and the maintenance of the pro-proliferative setting needed for proper skeletogenesis (Dentice et al., 2005). This interplay between WSB-1 and D2 could also be of significant physiological significance for the mammalian brain, as WSB-1 is expressed in tanycytes and astrocytes; two high D2-expressing tissues (Fekete et al., 2007)
TEB4 (aka MARCH6) is an ER resident, RING-finger containing protein, with 13–14 membrane-spanning domains where it may contain also a Sec61-like re-translocation channel (Kreft et al., 2006, Hassink et al., 2005). Several studies in yeast show that TEB4 is part of the ERAD machinery, interacts with the UBC6 E2 conjugase, and is responsible for the degradation of MATα2 gene product in yeast (Swanson et al., 2001). More recently, studies searching for targets of Doa10-mediated degradation dependent on the E2 ligases UBC6/7 identified D2 as a possible substrate for Doa10 in yeast (Ravid et al., 2006). Subsequently, TEB4 was shown to be a D2-interacting protein in human cells, as both proteins are co-immunoprecipitated. Co-expression of increasing amounts of TEB4 and D2 decreases D2 protein levels and enzymatic activity. A complete cellular knock down of TEB4 via siRNA stabiliZes D2 protein/activity levels and impairs its ubiquitination induced by T4 (Zavacki et al., 2009). These data indicate that TEB4 is required for substrate-induced ubiquitination of D2, while also participating in the regulation of D2’s protein/activity half-life. Like WSB-1, TEB4 requires the C-terminal instability loop of D2 in order to interact and mediate D2 ubiquitination, as a truncated D2 molecule that lacks such motif is not sensitive to TEB4 knock down (Zavacki et al., 2009).
Noteworthy is that maximal cellular levels of D2 activity (~ 3-fold increase) are found when the proteasome is blocked by MG-132 treatment or when TEB4 is knocked down. Conversely, when WSB1 is knocked down, D2 activity rises only about 2-fold. This could be interpreted as indicating that WSB-1 mediated D2 ubiquitination does not necessarily activate the proteasomal pathway for D2 whereas TEB4 does. In fact, tissue expression analysis of WSB-1 and TEB4 indicates the existence of tissues in which expression of one ligase predominates over the other. For example, BAT expresses high levels of WSB-1 with little or no expression levels of TEB4 (Zavacki et al., 2009). This suggests that these ligases may have different contributions to D2 degradation and also that there are other possible regulatory mechanisms involved in D2 ubiquitination. Supporting this idea is that the thyroid gland has significant levels of D2 activity, but it does not express either WSB-1 or TEB4.
USP20 and USP33 rescue D2 from ubiquitination
The addition of ubiquitin moieties to target proteins is a reversible process. The enzymes responsible for catalyzing the cleavage of the removal of Ub chains are termed deubiquitinases (DUB) and are divided in five different classes; the ubiquitin C-terminal hydrolases (UCH), the ubiquitin-specific proteases (USP), the ovarian tumor proteases (OTU), the Josephins and the JAB1/MPN/MOV34 class of metalloenzymes (Komander et al., 2009, Komander, 2010). These enzymes can cleave the peptide bonds between two ubiquitin molecules in short or long free/substrate-bound ubiquitin chains (Komander et al., 2009, Komander, 2010). The fact that the human genome encodes ~79 DUB genes suggests these enzymes lack a certain degree of selectivity for ubiquitinated substrates. This is overcome by several post-transcriptional regulatory mechanisms like interactions with larger protein complex (i.e. the proteasome), which are able to modulate and direct the DUB activity to specific ubiquitin chains. Over the years, DUBs have been recogniZed as mediating a critical step in the ubiquitin-mediated signaling pathways, such as the control of NF-kB signaling (Komander et al., 2009, Harhaj and Dixit, 2011), ubiquitin homeostasis (Komander et al., 2009), apoptosis (Ramakrishna et al.,2011), HIF-1α signaling (Li et al., 2005), thyroid hormone signaling and adaptative thermogenesis (Curcio-Morelli et al., 2003b), and tumorigenesis (Yuasa-Kawada et al., 2009).
Out of the 5 classes of DUB, two enzymes termed USP33 (or VDU-1) and USP20 (or VDU-2) from the USP family of DUBs has been shown to interact with and regulate D2 protein levels and activity (Curcio-Morelli et al., 2003b). USP33 and USP20 were initially described as being interacting proteins as well as substrates of the pVHL-elonginCB-cullin-2 (VCB-CUL2) E3 ubiquitin ligase complex, which is structurally and functionally similar to SCF (Skp1/Cdc53/F-box) E3 ubiquitin ligase complex (Li et al., 2002b, Li et al., 2002a). USP33 and USP20 are ubiquitously expressed in human and mouse tissues, sharing a high degree (~ 59%) of sequence homology at the carboxyl and amino termini (Li et al., 2002b, Li et al., 2002a, Komander et al., 2009). On the carboxyl terminal of both USPs, lays the zinc finger ubiquitin-specific protease domain (ZnF-USP), which in other USPs has been shown to bind ubiquitin molecules (Komander et al., 2009, Komander, 2010). Conversely, recent data based on the crystal structure of USP33’s ZnF-USP domain suggests this domain in particular does not bind ubiquitin molecules and may have another function other then ubiquitin binding (Allen and Bycroft, 2007). Knowledge about interaction of D2 with both USP33 and USP20 was obtained via a yeast two-hybrid screen, where the COOH-terminal of D2 carboxyl was used as bait (Curcio-Morelli et al., 2003b). Both USPs interacted with this portion of the D2 molecule, which was also observed in mammalian cells by co-immunoprecipitation studies (Curcio-Morelli et al., 2003b). In accordance with its deubiquitinating role, co-expression of USP33/20 increases D2 activity and prolonges D2 protein half-life. This was accompanied by a decrease in the levels of ubiquitinated D2 molecules, thus confirming USP33/20 role in rescuing D2 from the proteasomal pathway (Curcio-Morelli et al., 2003b).
The physiological relevance of D2 deubiquitination was elucidated in the BAT, where USP33 is up regulated by cold-exposure, which coincided with a 26-fold increase in D2 activity. At the same time, Dio2 mRNA was increased “only” by a factor of 10, indicating a role for post-transcriptional regulation of D2 activity, such as deubiquitination, thus maximizing the cold-induced increase in D2 activity and local T3 production. Interestingly, the mRNA levels of the HIF-1α deubiquitinase USP20 were not elevated during cold exposure, indicating that there is a subtype-specificity contribution to the regulation of D2 activity/protein levels (Curcio-Morelli et al., 2003b). Besides BAT, USP33-mediated regulation of D2 can also take place in the brain, as USP33 and D2 expression is found in astrocytes and tanycytes, two subtypes of glial cells (Fekete et al., 2007).
Stressed out: D2 and thyroid hormone signaling under ER stress
Over the years, the ER was always described as an organelle responsible for intracellular Ca2+ storage and homeostasis, protein folding, glyco-sylation and export. Studies have now linked the ER with complex cellular responses to different environmental inputs, such as glucose homeostasis and insulin signaling (Ozcan et al., 2004, Ozcan et al., 2008, Ozcan et al., 2009, Park et al., 2010) and also neurodegenerative diseases (Lindholm et al., 2006), thus placing the ER as a mediator of various signaling cascades and cell functions (Schröder, 2008, Schröder and Sutcliffe, 2010). Any disturbance of ER functions that are not rapidly counterbalanced leads to accumulation of unfolded/misfolded proteins within the ER lumen, a condition known as ER stress. To cope with ER stress, cells trigger an evolutionary conserved ER-to-nucleus signaling cascade, that (i) stops global protein translation in order to decrease ER protein input, (ii) increases expression of chaperone proteins such as HSP40 and BiP, and (iii) accelerates and increases the input of misfolded proteins into the ERAD machinery (Schröder, 2008, Meusser et al., 2005, Vembar and Brodsky, 2008). The identification of TEB4 as a regulator of D2 and reports that TEB4 is part of the ERAD machinery induced by conditions that cause ER stress (Ravid et al., 2006, Swanson et al., 2001), prompted studies to seek whether D2 is a target of ERAD triggered by ER stress. Indeed, by exposing D2-expressing cells to different ER-stressing conditions, we observed that D2 activity was rapidly down regulated in as little as 1h, with significant increase in ER stress markers (Arrojo e Drigo, 2010). This down regulation was independent of transcriptional modulation of the Dio2 gene, as ER stress did not change Dio2 mRNA levels. This loss of D2 activity leads to a significant decrease in D2-mediated T3 production, which shows that D2-expressing cells under ER stress are hypothyroid (Arrojo e Drigo, 2010).
ER stress can be attenuated or reversed by treatment with a small class of molecules termed chemical chaperones. These molecules are able to bind and stabiliZe protein conformation (Engin and Hotamisligil, 2010). Two chemical chaperones, tauroursodeoxycholic acid (TUDCA) and 4-phenyl butyric acid (4-PBA) have been shown to have metabolic activity, and treatment with such molecules normaliZed glucose homeostasis in animals fed a high fat diet (Ozcan et al., 2006), restored leptin sensitivity in ob/ob background mice (Ozcan et al., 2009) and a daily oral dose of 4-PBA improved lipid-induced insulin resistance and β-cell function (Xiao et al., 2011). In addition, both TUDCA and 4-PBA are being tested as therapeutic treatment of several diseases, ranging from amyotrophic lateral sclerosis (ALS) to obesity and metabolic syndrome (For more detailed information, see http://clinicaltrials.gov/). Although these metabolic effects of TUDCA and 4-PBA are well characteriZed, little is known about the specific molecular/cellular mechanisms mediating such effects. The fact that D2 is regulated by proteins involved in the ER stress cellular response (i.e. UBC6 and TEB4) and that chemical chaperones reverse the adverse effects of ER stress and improve metabolic parameter, led us to test whether treatment with TUDCA and 4-PBA would also affect D2 protein/activity levels. Initially, a dose and time dependent treatment of endogenously D2-expressing cell lines with TUDCA or 4-PBA led to a 2–3-fold increase in D2 activity, which coincided with an increase in intracellular T3 production that ranged from 2–10-fold (da-Silva et al., 2011). One could speculate that since chemical chaperones act to stabiliZe protein conformation, D2 levels were increased due to a longer D2-protein half-life. In fact, D2 protein/activity half-life was not changed and the increase in D2 total levels is due to transcriptional induction of the Dio2 gene by both TUDCA and 4-PBA (da-Silva et al., 2011). Similar results were also observed on a more physiological setting, where in vitro differentiated primary brown adipocytes where treated with either TUDCA or 4-PBA. As a result, in TUDCA-treated cells D2 activity increased approximately 3-fold, whereas with 4-PBA treatment induced D2 activity over 5-fold. This TUDCA- or 4-PBA-induced increase in D2 activity led to a significant acceleration in cellular oxygen consumption, lasting up to 72h. Strikingly, the TUDCA- or 4-PBA-induced oxygen consumption was reduced or lost in primary brown adipocytes isolated from Dio2−/− mice, clearly indicating a D2-dependent effect on oxygen consumption (da-Silva et al., 2011).
In the same setting, TUDCA and 4-PBA also induced the expression of key metabolic genes, such as Glut-4, CPT-1, UCP-1/3 and PGC-1α. With the exception of Glut-4, the induction of these genes with TUDCA and 4-PBA was limited when Dio2−/− brown adipocyte cells were used, clearly indicating that D2-mediated T3 production is critical for a proper metabolic response to these two chemical chaperones (da-Silva et al., 2011). The importance of the interplay between chemical chaperones and the D2-generated T3 pathway is underscored by the observation that treatment of wild-type, but not Dio2−/−, mice fed a high fat diet with TUDCA (0.5mg/kg BW) for 7 days led to increased BAT D2 activity, normalization of glucose homeostasis and increased fat oxidation (da-Silva et al., 2011). Taken together, these observations centrally place the D2 pathway and the acceleration of T3 production as a key player in mediating the metabolic effects seen after treatment with TUDCA or 4-PBA (da-Silva et al., 2011).
Concluding remarks
Thyroid hormone signaling can be tailored in a time and cell specific manner by the deiodinases, thus ensuring proper T3 effects in both developing and adult tissues. D2 drives thyroid hormone activation and cellular metabolism, and its activity is tightly regulated by an on/off switch mechanism triggered by substrate catalysis that involves enzyme inactivation via ubiquitination and reactivation via deubiquitination. D2 expression in metabolically relevant tissues such as BAT and skeletal muscle can be induced by bile acids, kaempferol and chemical chaperones, accelerating the rate of energy expenditure. This elevates D2 to a potential target of pharmacological intervention and treatment of the metabolic disorders.
Figure 1.
Thyroid hormone action is modulated by the deiodinases. T3 generated via D2-mediated deiodination acts via non-genomic mechanisms or the canonical nuclear thyroid hormone receptor pathway to regulate TSH and TRH secretion as well as cell development and metabolism. D3 terminates the thyroid hormone signaling pathway by inactivating T3 molecules into inactive substrates rT3 and/or T2.
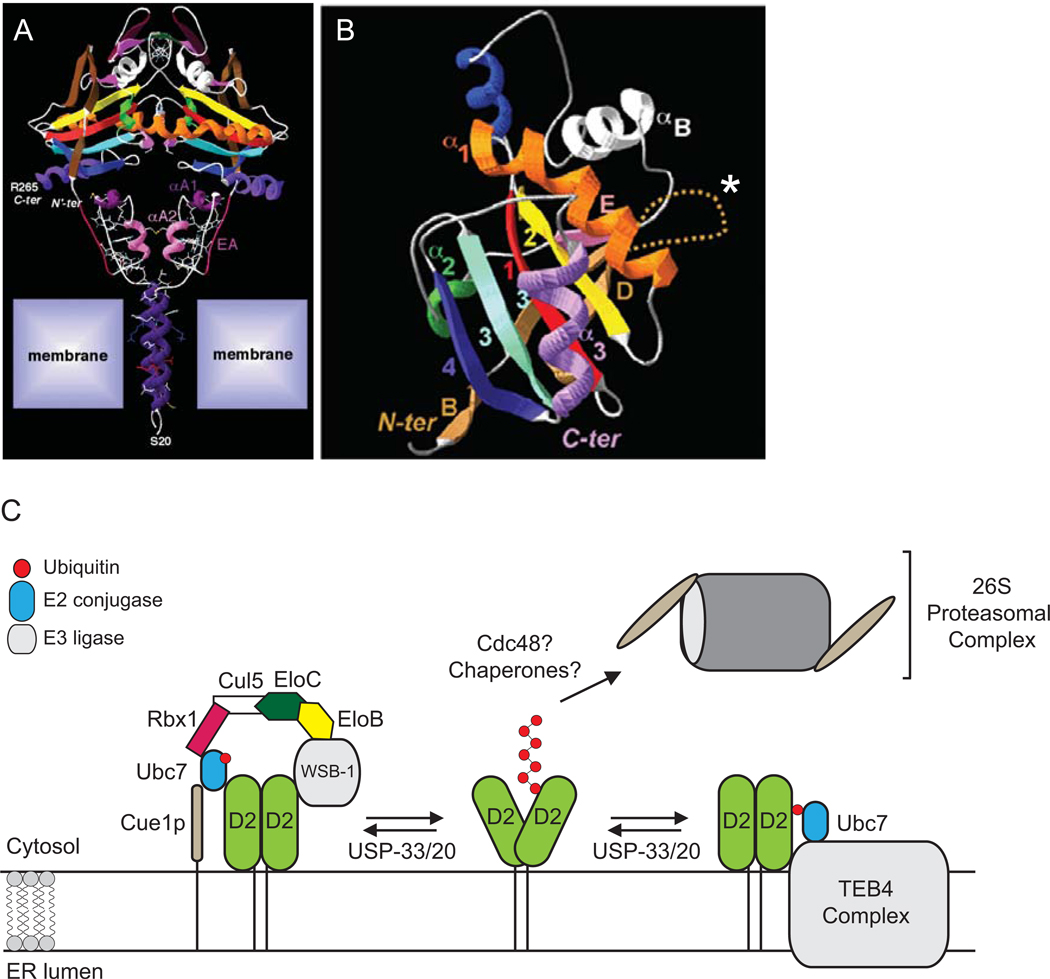
Figure 2.
D2 is a dimeric selenoenzyme ubiquitinated by WSB-1 and TEB4 complexes. (A) Ultra-structure of the D2:D2 dimer inserted into the ER membrane. Reproduced from (Sagar et al., 2007). (B) Detailed structure of the globular domain of D2. White asterisk marks the instability loop. Modified from (Dentice et al., 2005). In (C), newly synthesiZed D2 is targeted to the ER membrane, where D2 ubiquitination is triggered by T4-to-T3 catalysis. Ubiquitinated D2 (Ub-D2) is shuttled to the proteasomes. D2 can be rescued from terminal destruction via two DUBs: ubiquitin-specific proteases USP-20 and USP-33.
Table 1.
Tissue distribution of deiodinases in humans
| Deiodinase | Tissue |
|---|---|
| D1 | liver, thyroid, kidney |
| D2 | brain, pituitary gland, BAT, thyroid, muscle |
| D3 | developing tissues and placenta, adult skin, brain |
Acknowledgement
The authors have NIDDK support through DK58538, DK65055 and DK78620.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Allen MD, Bycroft M. The solution structure of the ZnF UBP domain of USP33/VDU1. Protein Sci. 2007;16:2072–2075. doi: 10.1110/ps.072967807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai R, Yoshikawa S, Murayama K, Imai Y, Takahashi R, Shirouzu M, Yokoyama S. Structure of human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7) Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:330–334. doi: 10.1107/S1744309106009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archange C, Nowak J, Garcia S, Moutardier V, Calvo EL, Dagorn JC, Iovanna JL. The WSB1 gene is involved in pancreatic cancer progression. PLoS One. 2008;3:e2475. doi: 10.1371/journal.pone.0002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrojo e Drigo R. International Thyroid Congress (ITC) Paris: 2010. Endoplasmatic Reticulum Stress and Chemical Chaperones regulate Type 2 Deiodinase and Thyroid Hormone Signaling. [Google Scholar]
- Baqui M, Botero D, Gereben B, Curcio C, Harney JW, Salvatore D, Sorimachi K, Larsen PR, Bianco AC. Human type 3 iodothyronine selenodeiodinase is located in the plasma membrane and undergoes rapid internalization to endosomes. J Biol Chem. 2003;278:1206–1211. doi: 10.1074/jbc.M210266200. [DOI] [PubMed] [Google Scholar]
- Baqui MM, Gereben B, Harney JW, Larsen PR, Bianco AC. Distinct subcellular localization of transiently expressed types 1 and 2 iodothyronine deiodinases as determined by immunofluorescence confocal microscopy. Endocrinology. 2000;141:4309–4312. doi: 10.1210/endo.141.11.7872. [DOI] [PubMed] [Google Scholar]
- Bassett JH, Boyde A, Howell PG, Bassett RH, Galliford TM, Archanco M, Evans H, Lawson MA, Croucher P, St Germain DL, Galton VA, Williams GR. Optimal bone strength and mineralization requires the type 2 iodothyronine deiodinase in osteoblasts. Proc Natl Acad Sci U S A. 2010;107:7604–7609. doi: 10.1073/pnas.0911346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283:12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Larsen PR. Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature. 1991;349:438–440. doi: 10.1038/349438a0. [DOI] [PubMed] [Google Scholar]
- Bianco AC, Kieffer JD, Silva JE. Adenosine 3',5'-monophosphate and thyroid hormone control of uncoupling protein messenger ribonucleic acid in freshly dispersed brown adipocytes. Endocrinology. 1992;130:2625–2633. doi: 10.1210/endo.130.5.1374009. [DOI] [PubMed] [Google Scholar]
- Bianco AC, Kim BW. Deiodinases: implications of the local control of thyroid hormone action. J Clin Invest. 2006;116:2571–2579. doi: 10.1172/JCI29812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev. 2002;23:38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- Botero D, Gereben B, Goncalves C, De Jesus LA, Harney JW, Bianco AC. Ubc6p and ubc7p are required for normal and substrate-induced endoplasmic reticulum-associated degradation of the human selenoprotein type 2 iodothyronine monodeiodinase. Mol Endocrinol. 2002;16:1999–2007. doi: 10.1210/me.2002-0135. [DOI] [PubMed] [Google Scholar]
- Briggman KB, Majumdar A, Coleman CS, Chau V, Tolman JR. NMR assignment of human ubiquitin conjugating enzyme Ubc7. J Biomol NMR. 2005;32:340. doi: 10.1007/s10858-005-1257-7. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Curcio-Morelli C, Mornon JP, Gereben B, Buettner C, Huang S, Castro B, Fonseca TL, Harney JW, Larsen PR, Bianco AC. The iodothyronine selenodeiodinases are thioredoxin-fold family proteins containing a glycoside hydrolase clan GH-A-like structure. J Biol Chem. 2003;278:36887–36896. doi: 10.1074/jbc.M305725200. [DOI] [PubMed] [Google Scholar]
- Canani LH, Capp C, Dora JM, Meyer EL, Wagner MS, Harney JW, Larsen PR, Gross JL, Bianco AC, Maia AL. The type 2 deiodinase A/G (Thr92Ala) polymorphism is associated with decreased enzyme velocity and increased insulin resistance in patients with type 2 diabetes mellitus. J Clin Endocrinol Metab. 2005;90:3472–3478. doi: 10.1210/jc.2004-1977. [DOI] [PubMed] [Google Scholar]
- Capelo LP, Beber EH, Huang SA, Zorn TM, Bianco AC, Gouveia CH. Deiodinase-mediated thyroid hormone inactivation minimizes thyroid hormone signaling in the early development of fetal skeleton. Bone. 2008;43:921–930. doi: 10.1016/j.bone.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho SD, Kimura ET, Bianco AC, Silva JE. Central role of brown adipose tissue thyroxine 5'-deiodinase on thyroid hormone-dependent thermogenic response to cold. Endocrinology. 1991;128:2149–2159. doi: 10.1210/endo-128-4-2149. [DOI] [PubMed] [Google Scholar]
- Castillo M, Hall JA, Correa-Medina M, Ueta C, Won Kang H, Cohen DE, Bianco AC. Disruption of Thyroid Hormone Activation in Type 2 Deiodinase Knockout Mice Causes Obesity With Glucose Intolerance and Liver Steatosis Only at Thermoneutrality. Diabetes. 2011 doi: 10.2337/db10-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QR, Bilke S, Wei JS, Greer BT, Steinberg SM, Westermann F, Schwab M, Khan J. Increased WSB1 copy number correlates with its over-expression which associates with increased survival in neuroblastoma. Genes Chromosomes Cancer. 2006;45:856–862. doi: 10.1002/gcc.20349. [DOI] [PubMed] [Google Scholar]
- Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–170. doi: 10.1210/er.2009-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiamolera MI, Wondisford FE. Minireview: Thyrotropin-releasing hormone and the thyroid hormone feedback mechanism. Endocrinology. 2009;150:1091–1096. doi: 10.1210/en.2008-1795. [DOI] [PubMed] [Google Scholar]
- Choi DW, Seo YM, Kim EA, Sung KS, Ahn JW, Park SJ, Lee SR, Choi CY. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J Biol Chem. 2008;283:4682–4689. doi: 10.1074/jbc.M708873200. [DOI] [PubMed] [Google Scholar]
- Christoffolete MA, Ribeiro R, Singru P, Fekete C, da Silva WS, Gordon DF, Huang SA, Crescenzi A, Harney JW, Ridgway EC, Larsen PR, Lechan RM, Bianco AC. Atypical expression of type 2 iodothyronine deiodinase in thyrotrophs explains the thyroxine-mediated pituitary thyrotropin feedback mechanism. Endocrinology. 2006;147:1735–1743. doi: 10.1210/en.2005-1300. [DOI] [PubMed] [Google Scholar]
- Cook WJ, Martin PD, Edwards BF, Yamazaki RK, Chau V. Crystal structure of a class I ubiquitin conjugating enzyme (Ubc7) from Saccharomyces cerevisiae at 2.9 angstroms resolution. Biochemistry. 1997;36:1621–1627. doi: 10.1021/bi962639e. [DOI] [PubMed] [Google Scholar]
- Curcio-Morelli C, Gereben B, Zavacki AM, Kim BW, Huang S, Harney JW, Larsen PR, Bianco AC. In vivo dimerization of types 1, 2, and 3 iodothyronine selenodeiodinases. Endocrinology. 2003a;144:937–946. doi: 10.1210/en.2002-220960. [DOI] [PubMed] [Google Scholar]
- Curcio-Morelli C, Zavacki AM, Christofollete M, Gereben B, de Freitas BC, Harney JW, Li Z, Wu G, Bianco AC. Deubiquitination of type 2 iodothyronine deiodinase by von Hippel-Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. J Clin Invest. 2003b;112:189–196. doi: 10.1172/JCI18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da-Silva WS, Harney JW, Kim BW, Li J, Bianco SD, Crescenzi A, Christoffolete MA, Huang SA, Bianco AC. The small polyphenolic molecule kaempferol increases cellular energy expenditure and thyroid hormone activation. Diabetes. 2007;56:767–776. doi: 10.2337/db06-1488. [DOI] [PubMed] [Google Scholar]
- da-Silva WS, Ribich S, Arrojo e Drigo R, Castillo M, Patti ME, Bianco AC. The chemical chaperones tauroursodeoxycholic and 4-phenylbutyric acid accelerate thyroid hormone activation and energy expenditure. FEBS Lett. 2011;585:539–544. doi: 10.1016/j.febslet.2010.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus LA, Carvalho SD, Ribeiro MO, Schneider M, Kim SW, Harney JW, Larsen PR, Bianco AC. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J Clin Invest. 2001;108:1379–1385. doi: 10.1172/JCI13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentice M, Bandyopadhyay A, Gereben B, Callebaut I, Christoffolete MA, Kim BW, Nissim S, Mornon JP, Zavacki AM, Zeöld A, Capelo LP, Curcio-Morelli C, Ribeiro R, Harney JW, Tabin CJ, Bianco AC. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat Cell Biol. 2005;7:698–705. doi: 10.1038/ncb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentice M, Marsili A, Ambrosio R, Guardiola O, Sibilio A, Paik JH, Minchiotti G, DePinho RA, Fenzi G, Larsen PR, Salvatore D. The FoxO3/type 2 deiodinase pathway is required for normal mouse myogenesis and muscle regeneration. J Clin Invest. 2010;120:4021–4030. doi: 10.1172/JCI43670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentice M, Salvatore D. Local impact of thyroid hormone inactivation. J Endocrinol. 2011 doi: 10.1530/JOE-11-0002. [DOI] [PubMed] [Google Scholar]
- Dohán O, De la Vieja A, Paroder V, Riedel C, Artani M, Reed M, Ginter CS, Carrasco N. The sodium/iodide Symporter (NIS): characterization, regulation, and medical significance. Endocr Rev. 2003;24:48–77. doi: 10.1210/er.2001-0029. [DOI] [PubMed] [Google Scholar]
- Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat Genet. 2005;37:1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- Dumitrescu AM, Liao XH, Best TB, Brockmann K, Refetoff S. A novel syndrome combining thyroid and neurological abnormalities is associated with mutations in a monocarboxylate transporter gene. Am J Hum Genet. 2004;74:168–175. doi: 10.1086/380999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Hotamisligil GS. Restoring endoplasmic reticulum function by chemical chaperones: an emerging therapeutic approach for metabolic diseases. Diabetes Obes Metab. 2010;12 Suppl 2:108–115. doi: 10.1111/j.1463-1326.2010.01282.x. [DOI] [PubMed] [Google Scholar]
- Fekete C, Freitas BC, Zeöld A, Wittmann G, Kádár A, Liposits Z, Christoffolete MA, Singru P, Lechan RM, Bianco AC, Gereben B. Expression patterns of WSB-1 and USP-33 underlie cell-specific posttranslational control of type 2 deiodinase in the rat brain. Endocrinology. 2007;148:4865–4874. doi: 10.1210/en.2007-0448. [DOI] [PubMed] [Google Scholar]
- Fekete C, Gereben B, Doleschall M, Harney JW, Dora JM, Bianco AC, Sarkar S, Liposits Z, Rand W, Emerson C, Kacskovics I, Larsen PR, Lechan RM. Lipopolysaccharide induces type 2 iodothyronine deiodinase in the mediobasal hypothalamus: implications for the nonthyroidal illness syndrome. Endocrinology. 2004;145:1649–1655. doi: 10.1210/en.2003-1439. [DOI] [PubMed] [Google Scholar]
- Freitas BC, Gereben B, Castillo M, Kalló I, Zeöld A, Egri P, Liposits Z, Zavacki AM, Maciel RM, Jo S, Singru P, Sanchez E, Lechan RM, Bianco AC. Paracrine signaling by glial cell-derived triiodothyronine activates neuronal gene expression in the rodent brain and human cells. J Clin Invest. 2010;120:2206–2217. doi: 10.1172/JCI41977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesema EC, Kuiper GG, Jansen J, Visser TJ, Kester MH. Thyroid hormone transport by the human monocarboxylate transporter 8 and its rate-limiting role in intracellular metabolism. Mol Endocrinol. 2006;20:2761–2772. doi: 10.1210/me.2005-0256. [DOI] [PubMed] [Google Scholar]
- Galton VA. The role of thyroid hormone in amphibian metamorphosis. Trends Endocrinol Metab. 1992;3:96–100. doi: 10.1016/1043-2760(92)90020-2. [DOI] [PubMed] [Google Scholar]
- Galton VA. The roles of the iodothyronine deiodinases in mammalian development. Thyroid. 2005;15:823–834. doi: 10.1089/thy.2005.15.823. [DOI] [PubMed] [Google Scholar]
- Gereben B, Goncalves C, Harney JW, Larsen PR, Bianco AC. Selective proteolysis of human type 2 deiodinase: a novel ubiquitin-proteasomal mediated mechanism for regulation of hormone activation. Mol Endocrinol. 2000;14:1697–1708. doi: 10.1210/mend.14.11.0558. [DOI] [PubMed] [Google Scholar]
- Gereben B, Zavacki AM, Ribich S, Kim BW, Huang SA, Simonides WS, Zeöld A, Bianco AC. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr Rev. 2008;29:898–938. doi: 10.1210/er.2008-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia CH, Christoffolete MA, Zaitune CR, Dora JM, Harney JW, Maia AL, Bianco AC. Type 2 iodothyronine selenodeiodinase is expressed throughout the mouse skeleton and in the MC3T3-E1 mouse osteoblastic cell line during differentiation. Endocrinology. 2005;146:195–200. doi: 10.1210/en.2004-1043. [DOI] [PubMed] [Google Scholar]
- Grozovsky R, Ribich S, Rosene ML, Mulcahey MA, Huang SA, Patti ME, Bianco AC, Kim BW. Type 2 deiodinase expression is induced by peroxisomal proliferator-activated receptor-gamma agonists in skeletal myocytes. Endocrinology. 2009;150:1976–1983. doi: 10.1210/en.2008-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Ribich S, Christoffolete MA, Simovic G, Correa-Medina M, Patti ME, Bianco AC. Absence of thyroid hormone activation during development underlies a permanent defect in adaptive thermogenesis. Endocrinology. 2010;151:4573–4582. doi: 10.1210/en.2010-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt B, Bause E. Lysine can be replaced by histidine but not by arginine as the ER retrieval motif for type I membrane proteins. Biochem Biophys Res Commun. 2002;291:751–757. doi: 10.1006/bbrc.2002.6515. [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Dixit VM. Deubiquitinases in the regulation of NF-κB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassink G, Kikkert M, van Voorden S, Lee SJ, Spaapen R, van Laar T, Coleman CS, Bartee E, Früh K, Chau V, Wiertz E. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J. 2005;388:647–655. doi: 10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemstra KA, Hoftijzer H, van der Deure WM, Peeters RP, Hamdy NA, Pereira A, Corssmit EP, Romijn JA, Visser TJ, Smit JW. The type 2 deiodinase Thr92Ala polymorphism is associated with increased bone turnover and decreased femoral neck bone mineral density. J Bone Miner Res. 2010;25:1385–1391. doi: 10.1002/jbmr.27. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Martinez ME, Fiering S, Galton VA, St Germain D. Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J Clin Invest. 2006;116:476–484. doi: 10.1172/JCI26240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TC, Lu X, Guo J, Wu JM. Differential regulation of proliferation, cell cycle control and gene expression in cultured human aortic and pulmonary artery endothelial cells by resveratrol. Int J Mol Med. 2010;26:743–749. doi: 10.3892/ijmm_00000521. [DOI] [PubMed] [Google Scholar]
- Huang SA, Bianco AC. Reawakened interest in type III iodothyronine deiodinase in critical illness and injury. Nat Clin Pract Endocrinol Metab. 2008;4:148–155. doi: 10.1038/ncpendmet0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Kim BW, Zavacki AM, Curcio-Morelli C, Dentice M, Harney JW, Larsen PR, Bianco AC. Endoplasmic reticulum-associated degradation of the human type 2 iodothyronine deiodinase (D2) is mediated via an association between mammalian UBC7 and the carboxyl region of D2. Mol Endocrinol. 2003;17:2603–2612. doi: 10.1210/me.2003-0082. [DOI] [PubMed] [Google Scholar]
- Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem. 2010;54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J Biol Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- Larsen PR. Thyroid-pituitary interaction: feedback regulation of thyrotropin secretion by thyroid hormones. N Engl J Med. 1982;306:23–32. doi: 10.1056/NEJM198201073060107. [DOI] [PubMed] [Google Scholar]
- Leonard JL, Siegrist-Kaiser CA, Zuckerman CJ. Regulation of type II iodothyronine 5'-deiodinase by thyroid hormone. Inhibition of actin polymerization blocks enzyme inactivation in cAMP-stimulated glial cells. J Biol Chem. 1990;265:940–946. [PubMed] [Google Scholar]
- Leonard JL, Visser TJ, Leonard DM. Characterization of the subunit structure of the catalytically active type I iodothyronine deiodinase. J Biol Chem. 2001;276:2600–2607. doi: 10.1074/jbc.M006973200. [DOI] [PubMed] [Google Scholar]
- Li Z, Na X, Wang D, Schoen SR, Messing EM, Wu G. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2002a;277:4656–4662. doi: 10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang D, Na X, Schoen SR, Messing EM, Wu G. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem Biophys Res Commun. 2002b;294:700–709. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Liu C, van Dyk D, Xu P, Choe V, Pan H, Peng J, Andrews B, Rao H. Ubiquitin chain elongation enzyme Ufd2 regulates a subset of Doa10 substrates. J Biol Chem. 2010;285:10265–10272. doi: 10.1074/jbc.M110.110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Ng L, Ma M, Kefas B, Davies TF, Hernandez A, Chan CC, Forrest D. Retarded developmental expression and patterning of retinal cone opsins in hypothyroid mice. Endocrinology. 2009;150:1536–1544. doi: 10.1210/en.2008-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Miura M, Tanaka K, Komatsu Y, Suda M, Yasoda A, Sakuma Y, Ozasa A, Nakao K. Thyroid hormones promote chondrocyte differentiation in mouse ATDC5 cells and stimulate endochondral ossification in fetal mouse tibias through iodothyronine deiodinases in the growth plate. J Bone Miner Res. 2002;17:443–454. doi: 10.1359/jbmr.2002.17.3.443. [DOI] [PubMed] [Google Scholar]
- Ng L, Goodyear RJ, Woods CA, Schneider MJ, Diamond E, Richardson GP, Kelley MW, Germain DL, Galton VA, Forrest D. Hearing loss and retarded cochlear development in mice lacking type 2 iodothyronine deiodinase. Proc Natl Acad Sci U S A. 2004;101:3474–3479. doi: 10.1073/pnas.0307402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Lyubarsky A, Nikonov SS, Ma M, Srinivas M, Kefas B, St Germain DL, Hernandez A, Pugh EN, Forrest D. Type 3 deiodinase, a thyroid-hormone-inactivating enzyme, controls survival and maturation of cone photoreceptors. J Neurosci. 2010;30:3347–3357. doi: 10.1523/JNEUROSCI.5267-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan L, Ergin AS, Lu A, Chung J, Sarkar S, Nie D, Myers MG, Ozcan U. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell Metab. 2009;9:35–51. doi: 10.1016/j.cmet.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Görgün C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Ozcan L, Yilmaz E, Düvel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Görgün CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Zhou Y, Lee J, Ozcan U. Sarco(endo)plasmic reticulum Ca2+-ATPase 2b is a major regulator of endoplasmic reticulum stress and glucose homeostasis in obesity. Proc Natl Acad Sci U S A. 2010;107:19320–19325. doi: 10.1073/pnas.1012044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol. 2010 doi: 10.1016/j.jhep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S, Suresh B, Baek KH. The role of deubiquitinating enzymes in apoptosis. Cell Mol Life Sci. 2011;68:15–26. doi: 10.1007/s00018-010-0504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosene ML, Wittmann G, Arrojo e Drigo R, Singru PS, Lechan RM, Bianco AC. Inhibition of the type 2 iodothyronine deiodinase underlies the elevated plasma TSH associated with amiodarone treatment. Endocrinology. 2010;151:5961–5970. doi: 10.1210/en.2010-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Leonard JL. Comparison of the physicochemical properties of type I and type II iodothyronine 5'-deiodinase. J Biol Chem. 1991;266:3233–3238. [PubMed] [Google Scholar]
- Sagar GD, Gereben B, Callebaut I, Mornon JP, Zeöld A, Curcio-Morelli C, Harney JW, Luongo C, Mulcahey MA, Larsen PR, Huang SA, Bianco AC. The thyroid hormone-inactivating deiodinase functions as a homodimer. Mol Endocrinol. 2008;22:1382–1393. doi: 10.1210/me.2007-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar GD, Gereben B, Callebaut I, Mornon JP, Zeöld A, da Silva WS, Luongo C, Dentice M, Tente SM, Freitas BC, Harney JW, Zavacki AM, Bianco AC. Ubiquitination-induced conformational change within the deiodinase dimer is a switch regulating enzyme activity. Mol Cell Biol. 2007;27:4774–4783. doi: 10.1128/MCB.00283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore D, Low SC, Berry M, Maia AL, Harney JW, Croteau W, St Germain DL, Larsen PR. Type 3 lodothyronine deiodinase: cloning, in vitro expression, and functional analysis of the placental selenoenzyme. J Clin Invest. 1995;96:2421–2430. doi: 10.1172/JCI118299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MJ, Fiering SN, Pallud SE, Parlow AF, St Germain DL, Galton VA. Targeted disruption of the type 2 selenodeiodinase gene (DIO2) results in a phenotype of pituitary resistance to T4. Mol Endocrinol. 2001;15:2137–2148. doi: 10.1210/mend.15.12.0740. [DOI] [PubMed] [Google Scholar]
- Schneider MJ, Fiering SN, Thai B, Wu SY, St Germain E, Parlow AF, St Germain DL, Galton VA. Targeted disruption of the type 1 selenodeiodinase gene (Dio1) results in marked changes in thyroid hormone economy in mice. Endocrinology. 2006;147:580–589. doi: 10.1210/en.2005-0739. [DOI] [PubMed] [Google Scholar]
- Schröder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65:862–894. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M, Sutcliffe L. Consequences of stress in the secretory pathway: The ER stress response and its role in the metabolic syndrome. Methods Mol Biol. 2010;648:43–62. doi: 10.1007/978-1-60761-756-3_3. [DOI] [PubMed] [Google Scholar]
- Silva JE. Physiological importance and control of non-shivering facultative thermogenesis. Front Biosci (Schol Ed) 2011;3:352–371. doi: 10.2741/s156. [DOI] [PubMed] [Google Scholar]
- Spang A. On vesicle formation and tethering in the ER-Golgi shuttle. Curr Opin Cell Biol. 2009;21:531–536. doi: 10.1016/j.ceb.2009.03.003. [DOI] [PubMed] [Google Scholar]
- St Germain DL, Galton VA. The deiodinase family of selenoproteins. Thyroid. 1997;7:655–668. doi: 10.1089/thy.1997.7.655. [DOI] [PubMed] [Google Scholar]
- St Germain DL, Galton VA, Hernandez A. Minireview: Defining the roles of the iodothyronine deiodinases: current concepts and challenges. Endocrinology. 2009;150:1097–1107. doi: 10.1210/en.2008-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Germain DL, Hernandez A, Schneider MJ, Galton VA. Insights into the role of deiodinases from studies of genetically modified animals. Thyroid. 2005;15:905–916. doi: 10.1089/thy.2005.15.905. [DOI] [PubMed] [Google Scholar]
- Steinsapir J, Bianco AC, Buettner C, Harney J, Larsen PR. Substrate-induced down-regulation of human type 2 deiodinase (hD2) is mediated through proteasomal degradation and requires interaction with the enzyme's active center. Endocrinology. 2000;141:1127–1135. doi: 10.1210/endo.141.3.7355. [DOI] [PubMed] [Google Scholar]
- Steinsapir J, Harney J, Larsen PR. Type 2 iodothyronine deiodinase in rat pituitary tumor cells is inactivated in proteasomes. J Clin Invest. 1998;102:1895–1899. doi: 10.1172/JCI4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Weissman AM. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s) J Biol Chem. 2001;276:16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Harpavat S, Billings NA, Cepko CL. Thyroid hormone components are expressed in three sequential waves during development of the chick retina. BMC Dev Biol. 2008;8:101. doi: 10.1186/1471-213X-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliauskas D, Hancock S, Stern CD. SWiP-1: novel SOCS box containing WD-protein regulated by signalling centres and by Shh during development. Mech Dev. 1999;82:79–94. doi: 10.1016/s0925-4773(99)00014-3. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser WE, Friesema EC, Visser TJ. Minireview: thyroid hormone transporters: the knowns and the unknowns. Mol Endocrinol. 2011;25:1–14. doi: 10.1210/me.2010-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- Xiao C, Giacca A, Lewis GF. Sodium Phenylbutyrate, a Drug With Known Capacity to Reduce Endoplasmic Reticulum Stress, Partially Alleviates Lipid-Induced Insulin Resistance and {beta}-Cell Dysfunction in Humans. Diabetes. 2011;60:918–924. doi: 10.2337/db10-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa-Kawada J, Kinoshita-Kawada M, Rao Y, Wu JY. Deubiquitinating enzyme USP33/VDU1 is required for Slit signaling in inhibiting breast cancer cell migration. Proc Natl Acad Sci U S A. 2009;106:14530–14535. doi: 10.1073/pnas.0801262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavacki AM, Arrojo E Drigo R, Freitas BC, Chung M, Harney JW, Egri P, Wittmann G, Fekete C, Gereben B, Bianco AC. The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol Cell Biol. 2009;29:5339–5347. doi: 10.1128/MCB.01498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavacki AM, Ying H, Christoffolete MA, Aerts G, So E, Harney JW, Cheng SY, Larsen PR, Bianco AC. Type 1 iodothyronine deiodinase is a sensitive marker of peripheral thyroid status in the mouse. Endocrinology. 2005;146:1568–1575. doi: 10.1210/en.2004-1392. [DOI] [PubMed] [Google Scholar]
- Zeöld A, Pormüller L, Dentice M, Harney JW, Curcio-Morelli C, Tente SM, Bianco AC, Gereben B. Metabolic instability of type 2 deiodinase is transferable to stable proteins independently of subcellular localization. J Biol Chem. 2006;281:31538–31543. doi: 10.1074/jbc.M604728200. [DOI] [PubMed] [Google Scholar]