Figure 4.
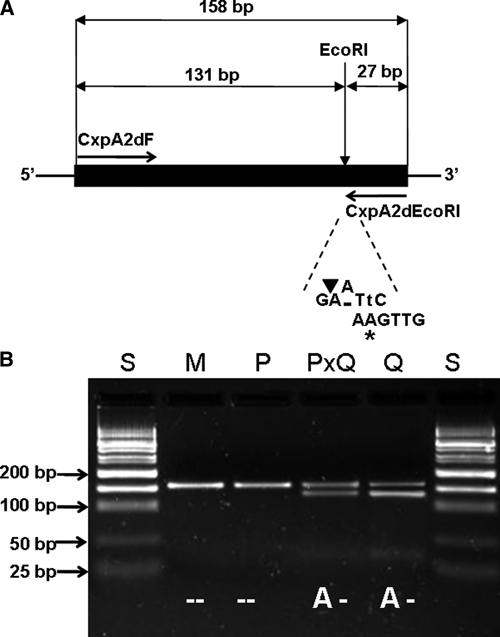
Design of CxpA2d SNP genotyping by PCR-EcoRI restriction assay. (A) Strategy of introducing mismatched PCR for A to deletion mutation. The nucleotide (A) marked with an asterisk indicates the mismatch (originally T) incorporated into the CxpA2dEcoRI primer to create a new EcoRI site (GAATTC) at the mutation location. The nucleotide in lower case indicates the complementary mismatch incorporated into the amplicon after PCR amplification. The solid black bar indicates the region corresponding to the amplicon. The black triangle indicates the cutting position of restriction enzyme EcoRI. (B) Electrophoretic patterns of digested PCR products amplified by primers CxpA2dF and CxpA2dEcoRI. S = 50 bp DNA size standard; M = pure Cx. p. pipiens f. molestus; P = pure Cx. p. pipiens f. pipiens; PXQ = hybrid of Cx. p. pipiens f. pipiens and Cx. p. quinquefasciatus; Q = Cx. p. quinquefasciatus.
