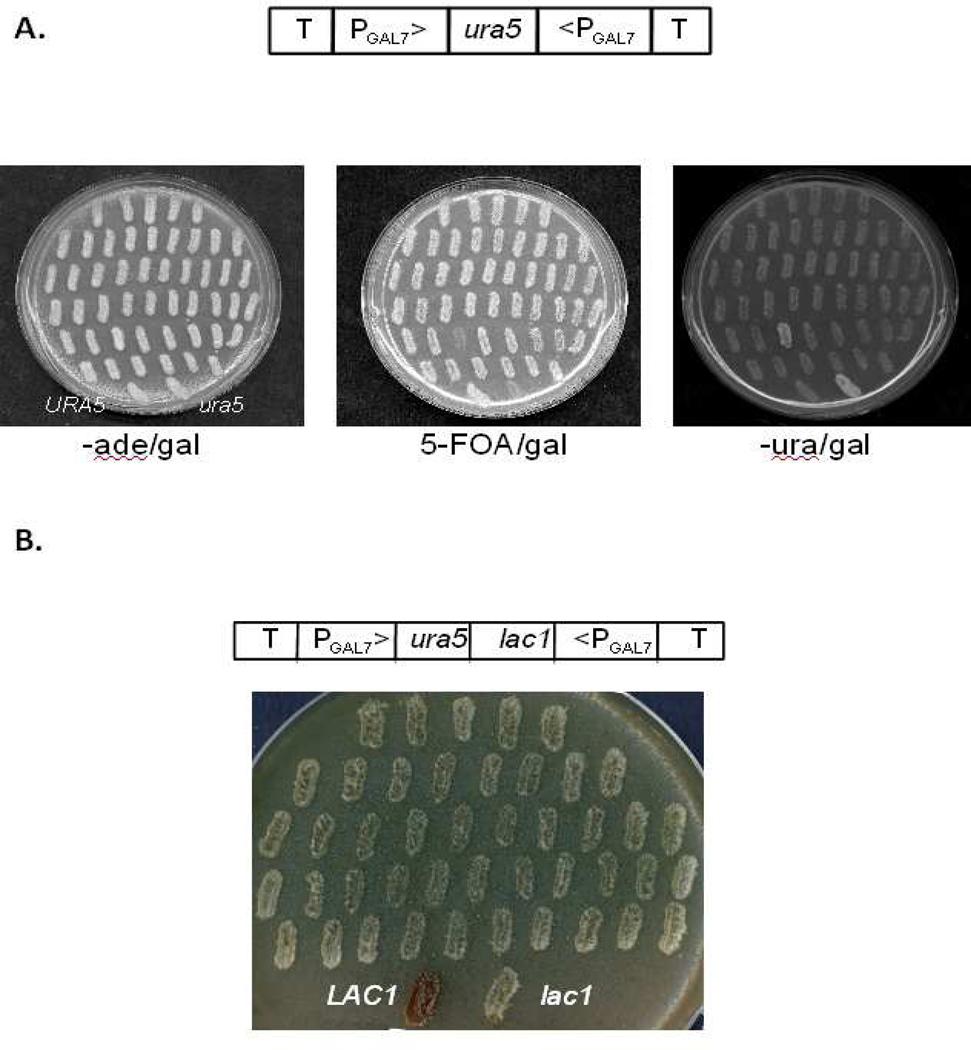
Figure 2.
Silencing URA5 allows 5-FOA selection of cells actively performing RNAi. A. Cells transformed with plasmid pIBB32 containing the RNAi cassette cartooned above were recovered and treated as described in the text before plating to medium with galactose but lacking adenine (left) and replica-plating to galactose media with 5-FOA (middle) or without uracil (right). Cells undergoing effective RNAi grow on 5-FOA and not on medium lacking uracil; this plate was chosen to show the single transformant (of over 100) that did not demonstrate this phenotype. The bottom two streaks on each plate are control strains with normal (left) or mutant (right) URA5 genes as indicated in the left panel. B. JEC50 cells were transformed with pIBB69 targeting LAC1 as cartooned above, pretreated with 5-FOA, and selected on –ade/gal medium as for the study in Fig. 2A. Transformants were restreaked on NSA/gal and grown for 2 days at 30°C, demonstrating effective silencing by white color matching that of control lac1 cells (bottom right of the plate). In contrast, the wild-type LAC1 strain is dark brown (bottom left).