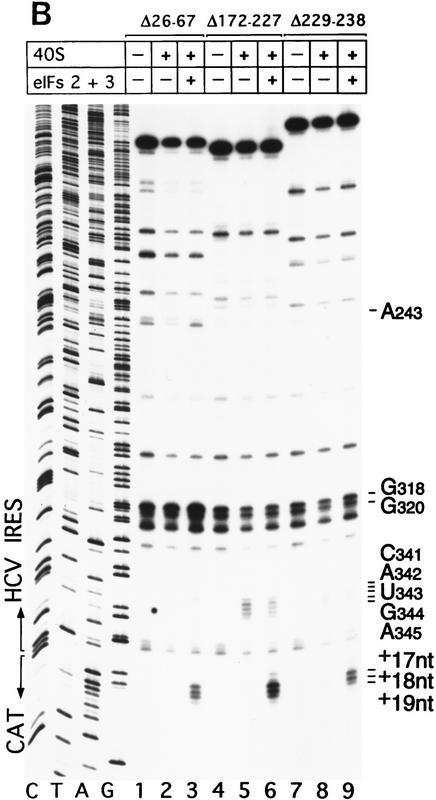
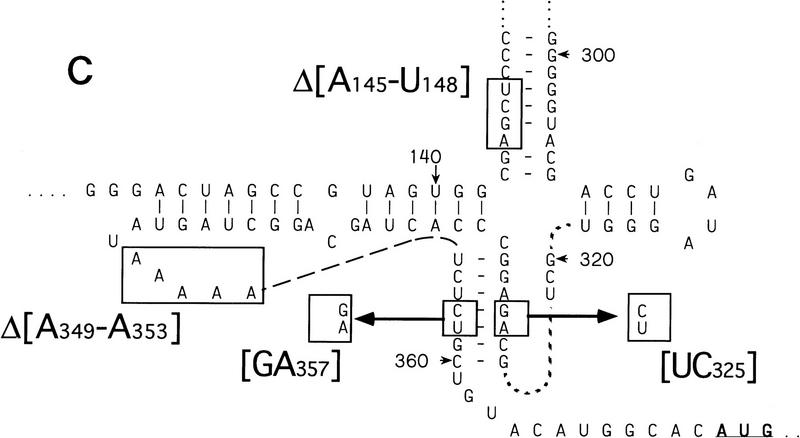
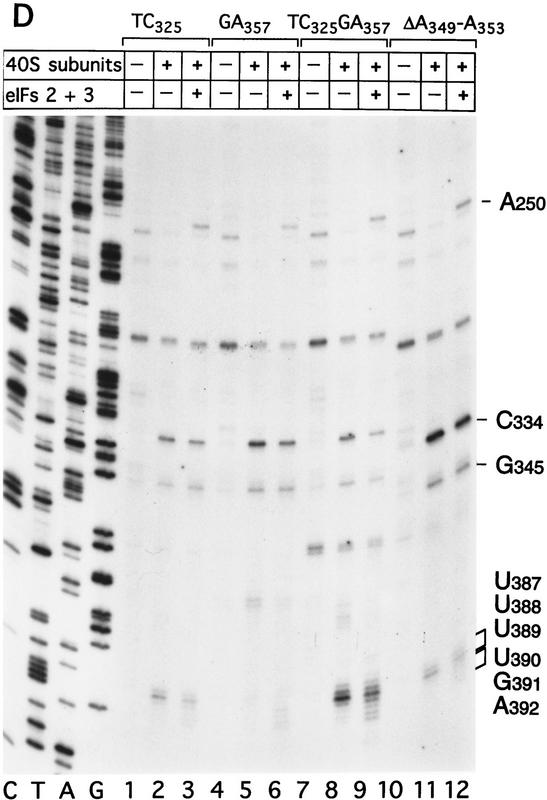
Figure 7.
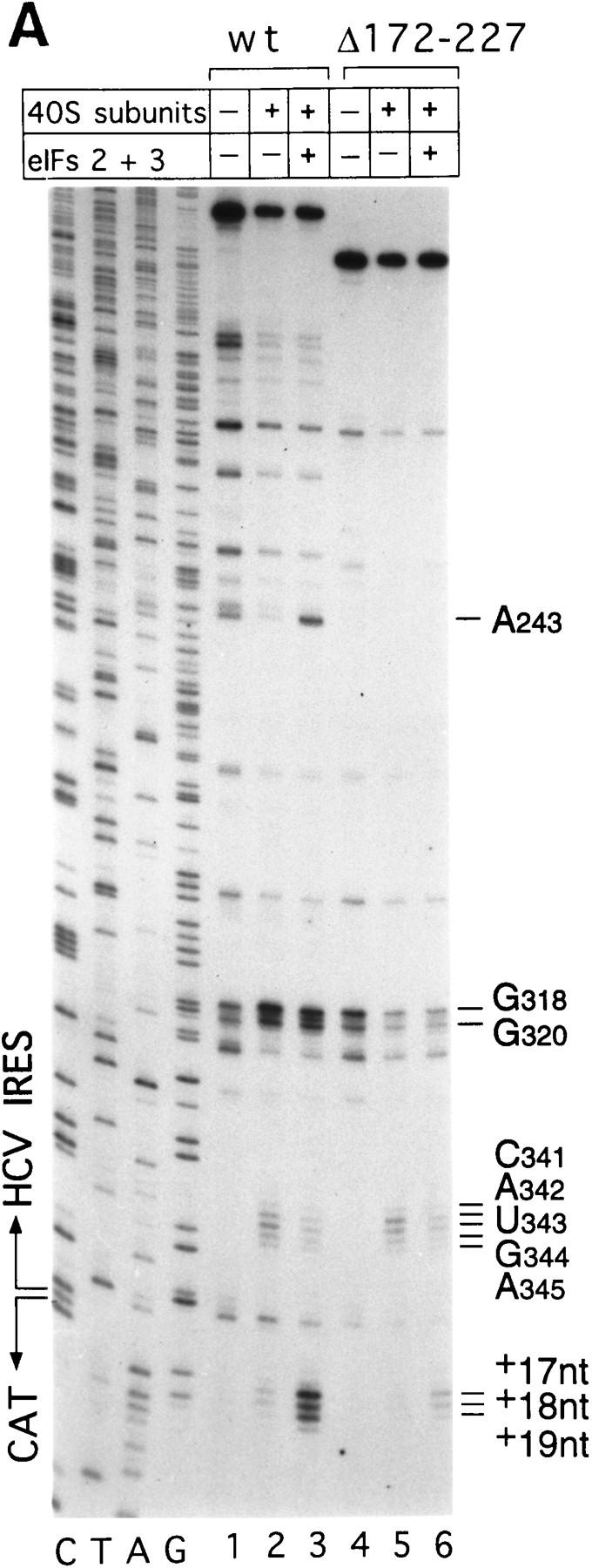
Binding of 40S subunits and assembly of 48S complexes on HCV and CSFV mutant IRESs. (A) Wild-type HCV (nucleotides 1–349)–CAT (lanes 1–3) and mutant HCV (nucleotides 1–349Δ172–227)–CAT (lanes 4–6) were incubated under standard conditions with 40S ribosomal subunits (lanes 2,3 and 5,6) and with eIF2, eIF3, Met–tRNAiMet and GMP–PNP (lanes 3,6). (B) Mutant HCV (nucleotides 1–380Δ26–67)–CAT (lanes 1–3), (nucleotides 1–380Δ172–227)–CAT (lanes 4–6) and (nucleotides 1–380Δ229–238) (lanes 7–9) RNAs were incubated with 40S subunits (lanes 2,3,5,6,8,9) and with eIF2, eIF3, Met–tRNAiMet, and GMP–PNP (lanes 3,6,9). Reaction conditions are described in Materials and Methods. A primer (5′-GCAACTGACTGAAATGCC-3′) was annealed to the CAT cistron ∼80 nucleotides downstream of the HCV initiation codon and was extended with AMV–RT. Primer extension was arrested at sites indicated on the right either as nucleotides in the HCV IRES or as positions relative to the A of the HCV initiation codon. Reference lanes C, T, A, and G depict the HCV–CAT sequence. The junction between the HCV IRES and the CAT cistron is indicated. This gel was exposed to x-ray film longer than that shown in A to show the differences in the intensity of stops on various mutant RNAs. (C) Model of the CSFV pseudoknot and adjacent nucleotides showing the substitutions present in the mutants UC325, GA357, and UC325GA357 and the deletions made in the mutants ΔA145–U148 and ΔA349–A353. In this model, helix I of the pseudoknot comprises nucleotides 129–142 and 331–346, and helix II comprises nucleotides 323–329 and 354–360. (D) CSFV (nucleotides 1–442)–NS′ RNAs containing the mutations UC325 (lanes 1–3), GA357 (lanes 4–6), UC325GA357 (lanes 7–9), and ΔA349–A353 (lanes 10–12) were incubated with 40S subunits (lanes 2,3,5,6,8,9,11,12) and with eIF2, eIF3, Met–tRNAiMet and GMP–PNP (lanes 3,6,9,12).