Figure 5.
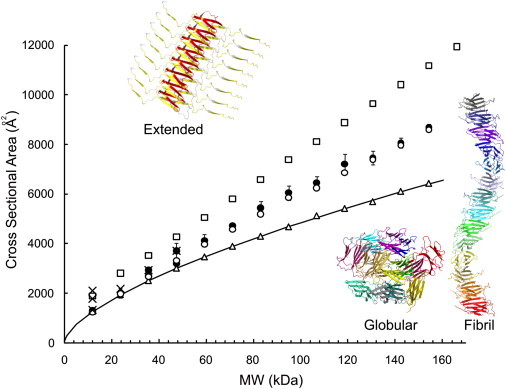
ESI-IMS-MS collision cross-sectional areas (Ω; Å2) for β2m oligomers detected during WL fibril assembly versus molecular mass (kDa). (Solid circles) β2m oligomers (dimer to 14-mer) populated during WL fibril assembly (100 mM ammonium acetate, pH 3.6). (Solid crosses) β2m oligomers (dimer to tetramer) populated during LS fibril assembly (100 mM ammonium formate, pH 2.5). (Error bars) One standard deviation in Ω between charge states of a protein. In silico models were constructed using the PyMOL Molecular Graphics System and their Ω-values calculated (34,40). In silico predicted Ω-values are shown for a linear assembly of nativelike β2m monomers stacked edge-to-edge (open circles), spherical oligomeric structures (open triangles), and an extended model with the N-terminal strand A and the C-terminal strand B of each protein subunit unstructured (open squares). (Solid line) The Ω expected for a protein of a given mass assuming a spherical assembly with a density of 0.44 Da Å−3 (31).
