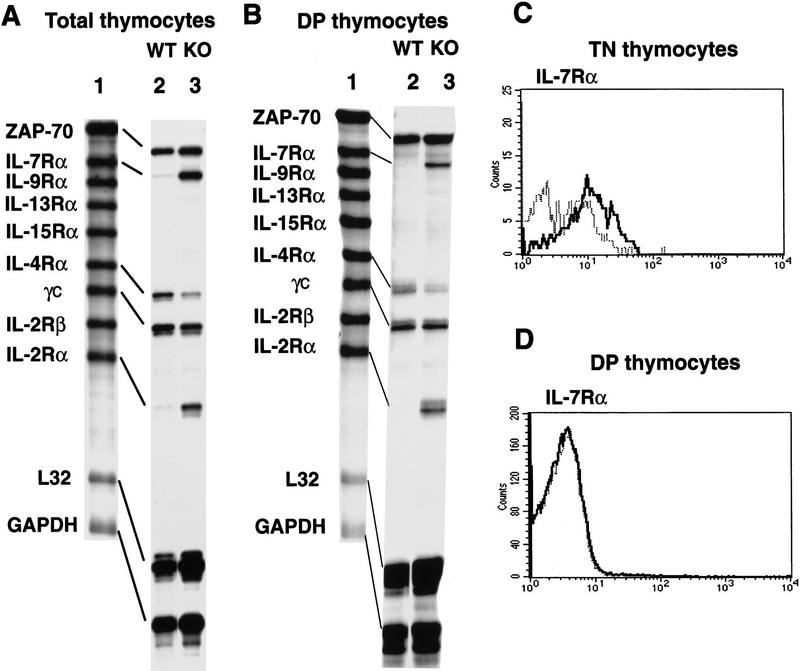
Figure 7.
Derepression of IL-2Rα and IL-7Rα genes in SATB1-null DP thymocytes. (A) RNA isolated from total thymocytes of SATB1-null and wild-type thymi were examined by a multi-probe RNase protection assay system containing a series of cytokine receptor probes. [32P]UTP-labeled antisense RNA probes (lane 1) were hybridized in excess to target total cellular RNA, after which free probe and other single-stranded RNAs were digested with RNase. The remaining RNase-protected probes derived from SATB1-null thymocytes (lane 2) and wild-type thymocytes (lane 3) were purified, and resolved on 6% denaturing polyacrylamide gels. (B) Identical to A except that target RNA was isolated from purified CD4+CD8+ DP cells by cell sorting using a PE-conjugated anti-CD8 antibody and an FITC-conjugated anti-CD4 antibody. (C) The CD3−CD4−CD8− TN cell subset from depleted thymocytes, as described in Fig. 3, were further purified by excluding Tri-Color-stained CD3-, CD4-, and CD8-positive T cells, B cells, granulocytes, dendritic cells, NK cells, and monocytes from thymocytes. The TN cell subset was subsequently subjected to FACS analysis with a PE-conjugated IL-7Rα antibody. (D) IL-7Rα+ cells in a CD4+CD8+ DP population were detected by a PE-conjugated anti-CD8 antibody and an FITC-conjugated anti-CD4 antibody with Cychrome–streptavidin combined with a biotin-conjugated anti-IL7Rα antibody. (C,D) (Bold line) SATB1 null mice; (narrow line) wild-type mice.