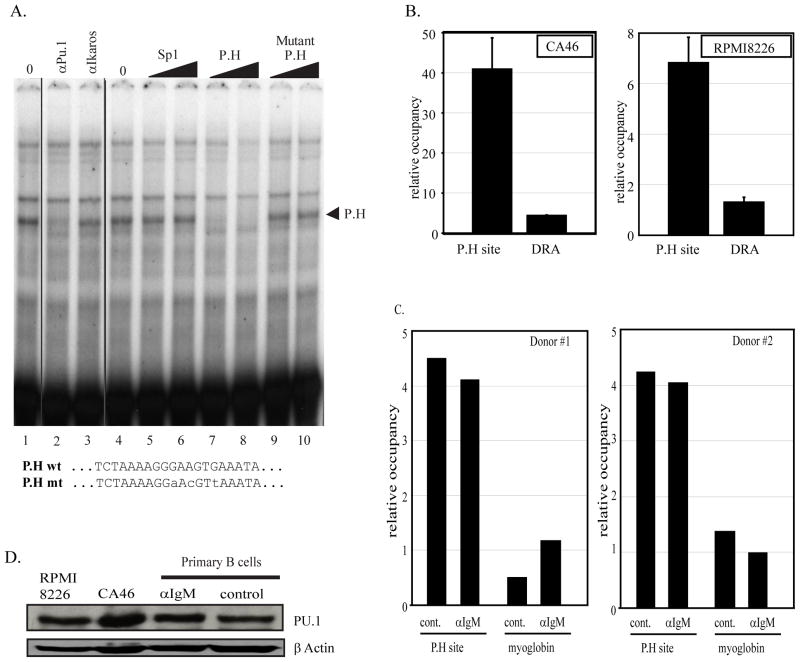
Figure 6. Site P.H is a PU.1 factor binding site.
A) In vitro binding assays were done using an oligonucleotide containing the P.H site sequence identified at position −1745 to −1737 of the PRDM1 promoter and CA46 nuclear extracts. Lanes 1 and 4 contain 2 uL of nuclear extract. Unlabeled competitor oligonucleotides as indicated at the top of lanes 5–10 were added to the binding reaction at 150 or 300-fold molar excess. A single complex indicated by an arrowhead is specifically competed by the wild type but not a mutant or unrelated oligonucleotide. In lanes 2 and 3 antibodies as indicated at the top were added to the binding reaction. The formation of the specific P.H complex is inhibited by addition of PU.1 antibody. The wild type and mutant P.H site sequences are shown at the bottom. B) Chromatin immunoprecipitation assay was performed in CA46 lymphoma cells (left panel) and RPMI8226 myeloma cells (right panel) using PU.1 antibody and quantitative PCR primers spanning the P.H site. PU.1 binding at the P.H. site was significantly higher than on the negative control promoter HLA-DRA. Data shown are mean of 3 independent experiments with SEM shown (p<0.01). C) Chromatin immunoprecipitation assay in activated primary human B cells. The experiment is as described in panel B except a myoglobin B locus is shown as the negative control. Lanes labeled αIgM were treated for 24 hours with anti-IgM while lanes labeled control were mock treated for 24 hours. PU.1 binding is specifically detected at the P.H site and is not altered by anti-IgM treatment. Similar data were obtained in two independent donor samples. D) Immunoblot analysis of PU.1 expression.