Abstract
Introduction
IKBKB (IKK-β/IKK-2), which activates NF-κB, is a substrate of the KEAP1-CUL3-RBX1 E3-ubiquitin ligase complex, implicating this complex in regulation of NF-κB signaling. We investigated complex component gene disruption as a novel genetic mechanism of NF-κB activation in non-small cell lung cancer (NSCLC).
Methods
644 tumor- and 90 cell line-genomes were analyzed for gene-dosage status of the individual complex components and IKBKB. Gene expression of these genes, and NF-κB target genes were analyzed in 48 tumors. IKBKB protein levels were assessed in tumors with and without complex or IKBKB genetic disruption. Complex component knockdown was performed to assess effects of the E3-ligase complex on IKBKB and NF-κB levels, and phenotypic importance of IKBKB expression was measured by pharmacological inhibition.
Results
We observed strikingly frequent genetic disruption (42%) and aberrant expression (63%) of the E3-ligase complex and IKBKB in the samples examined. While both adenocarcinomas and squamous cell carcinomas showed complex disruption, the patterns of gene disruption differed. IKBKB levels were elevated with complex disruption, knockdown of complex components increased activated forms of IKBKB and NF-κB proteins, and IKBKB inhibition detriments cell viability, highlighting the biological significance of complex disruption. NF-κB target genes were overexpressed in samples with complex disruption, further demonstrating the effect of complex disruption on NF-κB activity.
Conclusions
Gene dosage alteration is a prominent mechanism that disrupts each component of the KEAP1-CUL3-RBX1 complex and its NF-κB stimulating substrate, IKBKB. Here we show that, multiple component disruption of this complex represents a novel mechanism of NF-κB activation in NSCLC.
Keywords: KEAP1, CUL3, RBX1, IKBKB, NF-κB signaling, genetic disruption
Introduction
KEAP1 is a substrate adaptor protein that binds substrates to an E3-ubiquitin ligase complex which also includes CUL3 and RBX1 (Figure 1). Ubiquitination and degradation of complex substrates prevents their accumulation and undesired downstream effects. The best characterized substrate of KEAP1 and the E3-complex is NRF2 1–3. In response to oxidative stress, NRF2 stimulates transcription of cytoprotective genes that scavenge harmful reactive molecules, preventing cellular damage 4. Interestingly, lung specific Keap1 knockout in mice was shown to protect against cell damage caused by cigarette smoke by enabling Nrf2 accumulation and increased expression of its target genes 5. NRF2 has also been implicated in cancer cell resistance to chemotherapeutics by its activation of drug-metabolizing and drug-efflux proteins 4.
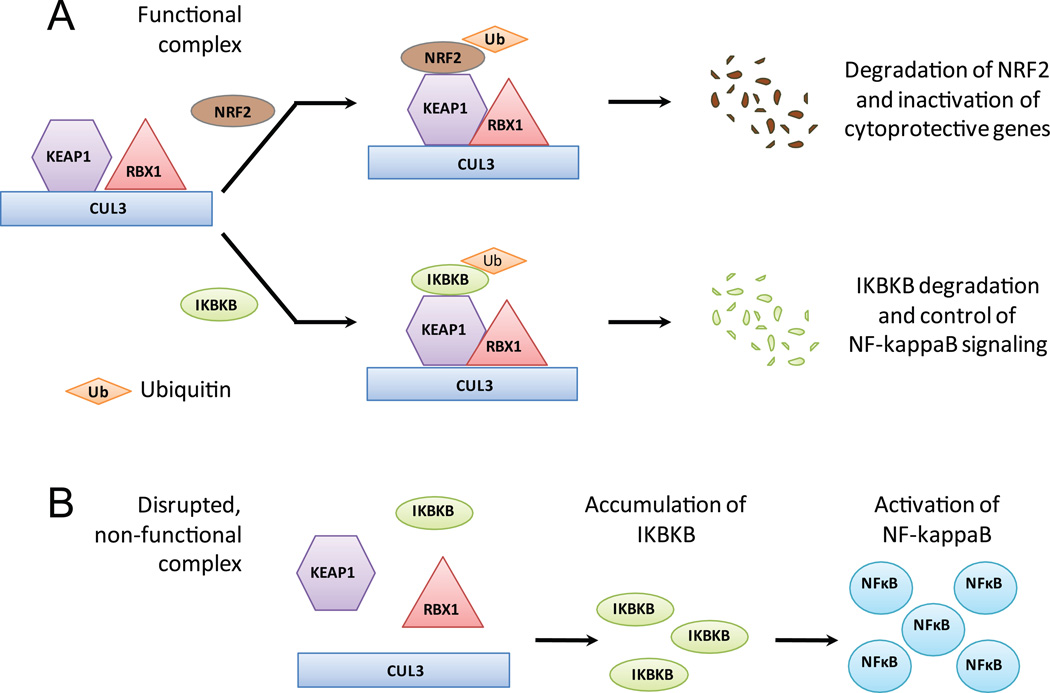
Figure 1.
The KEAP1 Cullin-3 E3-ubiquitin ligase complex and its roles in maintaining cellular levels of NRF2 and IKBKB. (A) When complex components are intact, KEAP1 facilitates binding of NRF2 or IKBKB which promotes their ubiquitination. This complex prevents accumulation of IKBKB and subsequent NF-κB activation. (B) Disruption of any complex component compromises function leading to stabilization and accumulation of IKBKB, and aberrant activation of NF-κB.
Recently, the KEAP1 E3-ligase complex has been implicated in the regulation of NF-κB signaling 6, which promotes cell proliferation and survival 7–9. The NF-κB pathway is activated in over 60% of lung cancers, however, the genetic mechanisms underlying its activation remain largely unknown 7, 10–13. When inactive, NF-κB is bound by inhibitory proteins (I-kappaB) in the cytoplasm. Upon stimulation, the kinase IKBKB phosphorylates IκB, releasing its inhibition, and enabling NF-κB translocation to the nucleus where it exerts its effects (Figure 1) 14. Lee et al. demonstrated that KEAP1 binds IKBKB, drawing it to the E3-ligase complex for ubiquitination and degradation, providing a mechanism of NF-κB regulation 6. These findings provide significant evidence of the integral role of KEAP1 and the E3-ligase complex in NF-κB signaling.
The ability of the E3-ligase complex to ubiquitinate IKBKB was most efficient when all three complex components were expressed and intact, suggesting disruption of even a single component compromises function. Although somatic DNA alterations have been observed in the genes encoding some of these complex components, it is not known if complex component gene disruptions are a key mechanism of NF-κB activation in lung cancer 6. In lung cancer, only KEAP1 has been thoroughly investigated at the DNA level, and a recent study reported low KEAP1 expression was associated with poor patient outcome 15. KEAP1 deletions, mutations and DNA hypermethylation have been reported in 3–41% of lung tumors 16–21, however, the moderate frequency of KEAP1 gene disruption alone is not sufficient to explain the high (>60%) frequency of NF-κB activation in lung cancer 11.
We propose that genetic disruption of any one E3-ubiquitin ligase complex component may be sufficient to result in tumorigenic NF-κB activation due to loss of complex function and subsequent accumulation of IKBKB 6, 14, 22. We hypothesize that somatic disruptions of CUL3 (2q36.2) and RBX1 (22q13.2), in addition to KEAP1 (19p13.2) occur frequently in lung tumors, representing a prominent genetic mechanism that may be responsible for IKBKB accumulation and stimulation of NF-κB. In this study, we 1) investigated whether these complex components and IKBKB (8p11.21) exhibit gene dosage and expression alterations and the frequencies at which they occur in a large cohort of clinical lung tumors, 2) investigated whether the complex components display subtype specific disruption, and 3) assessed the functional consequence of complex disruption on NF-κB activity, as these genetic events may be significant contributors to the NF-κB activation commonly observed in lung cancer.
Materials and Methods
Non-small cell lung cancer (NSCLC) samples
261 lung tumors (169 adenocarcinomas (AC) and 92 squamous cell carcinomas (SCC)) were accrued from Vancouver General Hospital (Vancouver, Canada) and Princess Margaret Hospital (Toronto, Canada) following ethics approval with patient consent (see Table, Supplemental Digital Content 1, which lists patient demographics). Tissue sections were microdissected with the guidance of lung pathologists. Matched non-malignant lung tissue was also obtained for a subset of the primary tumors collected. DNA for all 261 samples was extracted using standard phenol-chloroform procedures. RNA was extracted from tumor and matched non-malignant tissues using RNeasy Mini Kits (Qiagen Inc., Mississauga, ON). NSCLC cell lines (H1650, HCC827, H3255, H358, H23, HCC95, H2347, and H2122) were obtained from ATCC or the laboratory of AFG and cultivated as previously described 23. These cell lines were fingerprinted to confirm their identity 24. Human bronchial epithelial cells (HBEC-KT) were provided by Dr. John Minna (UTSW) and maintained as previously described 25. Primary non-malignant human bronchial epithelial (NHBE) lung cells were obtained from Lonza (Walkersville, MD).
Determination of gene dosage and expression levels
Copy number status (gain, loss, or neutral) for the KEAP1, CUL3, RBX1, and IKBKB loci was determined for each tumor sample and 63 NSCLC cell lines by array comparative genomic hybridization (array CGH) using the whole genome tiling path array (SMRT v.2, BCCRC Array Laboratory, Vancouver, BC) as previously described 26–28. Gene expression levels of KEAP1, CUL3, RBX1, and IKBKB were determined using custom Agilent gene expression microarrays in 35 AC and 13 SCC lung tumors and corresponding matched non-malignant tissues. Genes were classified as over or underexpressed if the fold change in mRNA expression levels in tumors relative to matched non-malignant tissues was greater or less than 2 fold. Gene expression for KEAP1, CUL3, RBX1, and IKBKB was also assessed in an additional, distinct cohort of 49 NSCLC (29 AC and 20 SCC) tumors with matched CGH profiles using custom Affymetrix arrays. The association between copy number and gene expression was assessed by segregating these 49 NSCLC tumors into those with and without copy number alterations for each gene as previously described 28. Expression levels were compared in both groups using a U test with a p-value less than 0.05 considered significant. The probe with the highest median intensity across all tumor samples was assessed for each gene.
Copy number analysis of external cohorts
Publically available NSCLC data was investigated to further explore the frequency of genomic disruption at the KEAP1, CUL3, RBX1, and IKBKB loci. Affymetrix SNP 250K data for 383 matched tumor non-malignant NSCLC pairs were accessed from the dbGaP Genotypes and Phenotypes database (Study Accession: phs000144.v1.p1). Partek Genomics Suite software was used to generate copy number profiles for all 383 tumors using each matched non-malignant sample as a baseline for defining copy number alterations. Affymetrix SNP 6.0 array profiles were obtained for 54 NSCLC cell lines, 27 of which overlapped with cell lines profiled on the SMRT array, from the Wellcome Trust Sanger Institute CGP Data Archive (sanger.ac.uk/genetics/CGP/Archive/) (see Table, Supplemental Digital Content 2, which lists NSCLC cell lines used in this study). Copy number profiles for these cell lines were also generated using Partek Genomics Suite software using profiles derived from SNP data for 72 cytogenetically normal HapMap individuals as a reference 29. In total, 90 unique NSCLC cell lines were analyzed and genetic disruption was noted if either platform (SNP or SMRT array) detected a copy number alteration. The Broad Institute's Tumorscape database (www.broadinstitute.org/tumorscape) was also accessed to investigate copy number status at these four gene loci 30.
Western blot assessment of total and phospho- IKBKB and total and phospho- NF-κB protein levels
IKBKB protein levels were assessed in 8 NSCLC cell lines with various combinations of KEAP1, CUL3, or RBX1 genomic loss or IKBKB genomic gain using antibodies from Cell Signaling (IKBKB #2678, p-IKBKB #2697, NF-κB: #4764, p-NF-κB: #3033, and GAPDH #2118 Beverly MA, USA). Western blots were performed following standard procedures as previously described 25. Cells were washed with cold PBS and lysed in RIPA buffer with complete protease inhibitor cocktail (Roche, Basel Switzerland). Protein lysates were quantified using the BCA assay (Fisher Scientific, Ottawa, Canada). Lysates were diluted and boiled for electrophoresis then transferred to a polyvinylidene membrane. Membranes were blocked in 5% skim milk or 5% BSA in Tris buffered saline containing Tween 20 (TBS-T) (according to the manufacturer's recommendations) and then incubated with primary antibody (1:1000) at 4°C overnight. Following three washes in TBS-T, membranes were incubated with HRP conjugated secondary antibody (Cell Signaling, cat. #7074, 1:20000) for 1 hour at room temperature. Antibody binding was visualized by enhanced chemiluminescence (Supersignal West Femto, Thermo Scientific, Rockford, IL, USA) after three washes in TBS-T.
Immunohistochemistry (IHC) staining for IKBKB protein levels
5 µm thick sections were cut from 13 formalin fixed, paraffin embedded tumor specimens with various states of genomic disruption to the E3-ubiquitin ligase complex genes or IKBKB. IHC to determine protein expression of IKBKB was performed as previously described 25. Briefly, slides were deparaffinized in xylene and rehydrated with graded ethanol washes. Antigen retrieval was performed using a decloaking chamber with sodium citrate buffer pH 6.0, after which endogenous peroxidase activity was blocked using 3% H2O2 for 30 minutes at room temperature. Sections were blocked with goat serum for 3 hours at room temperature and then incubated overnight at 4°C with 32 µg/ml of anti-IKBKB mouse monoclonal primary antibody (EMD4 Biosciences, cat. OP134, San Diego, CA, USA). Prior to incubation with an anti-mouse HRP-streptavidin conjugated secondary antibody (DAKO, cat. K4000, Glostrup, Denmark), four five minute washes in TBS-T were performed to remove unbound primary antibody. Detection of antibody binding was assessed using diaminobenzidine (Sigma Aldrich, cat. D4293, St. Louis, MO, USA). Slides were counterstained with hematoxylin for visualization. Intensity of staining was scored using a 0–3+ system based on the consensus of 3 observers (KT, LP, JCE). The mean staining intensity for each tumor section was judged as follows: 0 - no staining, 1 - weak intensity, 2 - moderate intensity, 3 - strong staining.
siRNA-mediated complex component knockdowns
On-Target plus SMART pool siRNAs targeting KEAP1, RBX1, CUL3, and a non-targeting control (NTC) pool of siRNAs were purchased from Dharmacon. One day prior to transfection, HBEC-KT cells were plated in regular growth media (antibiotic free) at a density of 200,000 cells/well in six well plates. Transfections were performed using an siRNA concentration of 100 nm according to the Thermo Scientific DharmaFECT siRNA transfection protocol. DharamaFECT 1 transfection reagent (Dharmacon) was used at a concentration of 0.2 µl/100 µl media. After 12 hours, transfection media was replaced with regular growth media. RNA was harvested from cells at 48 hours post transfection using the Trizol method (Invitrogen) and protein lysates were prepared using RIPA buffer 72 hours post transfection. Knockdown efficiencies were measured by qPCR with the following TaqMan assays from Applied Biosystems and using 18S as an endogenous control: Hs99999901_s1 (18S), Hs00202227_m1 (KEAP1), Hs00180183_m1 (CUL3), and Hs00360274_m1 (RBX1). Western blots were performed as above to measure total and phospho-IKBKB and NF-κB protein levels.
NF-κB target gene analysis
Nine genes transcriptionally controlled by NF-κB - as annotated in the Ingenuity Pathway Analysis database (Ingenuity® Systems, Redwood CA, USA) were analyzed (see Table, Supplemental Digital Content 3, which lists the nine NF-κB target genes analyzed). Wilcoxon signed-rank tests were used to compare NF-κB target gene expression in 48 NSCLC tumors with deregulated KEAP1, CUL3, RBX1 or IKBKB expression levels (2-fold or greater) versus that in their matched non-malignant lung tissues. NF-κB target genes were considered upregulated if expression levels were significantly elevated in tumors relative to matched non-malignant tissues (Wilcoxon p <0.05).
IKBKB inhibition in NSCLC cell lines
Cell viability assays were performed to measure the effect of IKBKB inhibition by a cell permeable, competitive ATP inhibitor, IKK-2 inhibitor IV (Calbiochem cat. #401481, San Diego, CA, USA) on five NSCLC cell lines (H3255, H2122, H23, HCC95 and H1650). Cells were plated in triplicate in 96 well plates at optimal densities for growth (H2122 and H23 at 2000 cells/well, H1650 and HCC95 at 3000 cells/well, and H3255 at 5000 cells/well). Cells were subjected to a series of 2-fold dilutions of IKBKB inhibitor which was prepared in cell growth media and DMSO. The experimental inhibitor concentrations ranged from 500 µM to 244 nM and the final DMSO concentration for treated and untreated (control) cells was 1%. Blank wells contained equal volumes of growth media with 1% DMSO. Cells were incubated for 72 hours at 37°C and then treated with 10µl of Alamar Blue cell viability reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. The reaction product was quantified by measuring absorbance at 570 nm with reference to 600 nm using an EMax plate reader (Molecular Devices, Sunnyvale, CA, USA). The average absorbance readings for blank wells were subtracted from all treatment and control wells and technical replicates were averaged. The response of treated cells was measured as a proportion of the viability of untreated cells, with the mean background subtracted treatment absorbance divided by the mean background subtracted untreated absorbance for each inhibitor concentration. Dose response curves and IC50 values were generated in Graph Pad v5 using the proportionate response of all 12 drug concentrations. Experiments were repeated in triplicate and differences in IC50 values were determined using a student's t-test with a p-value < 0.05 considered significant.
Results
Gene disruption of E3-ubiquitin ligase complex components in multiple NSCLC datasets
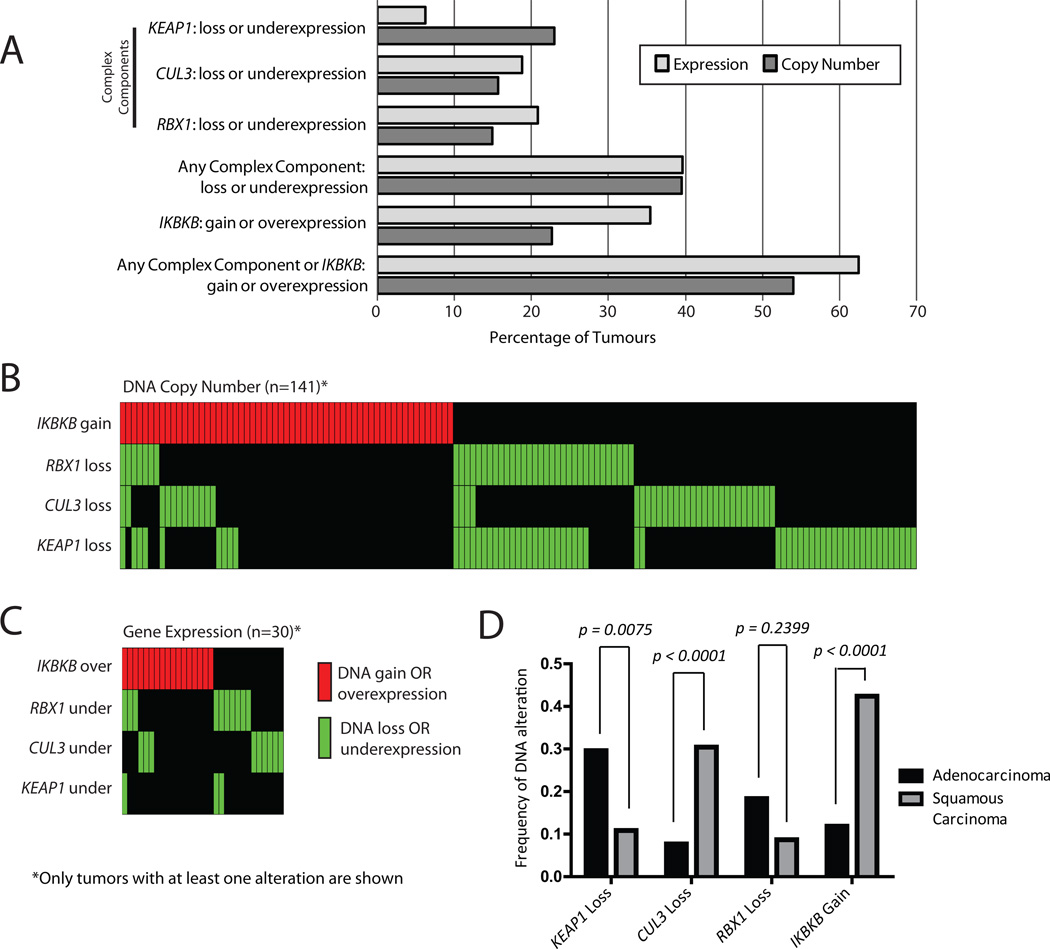
261 tumors were screened for DNA copy number alterations at the KEAP1, CUL3, and RBX1 loci. A significant proportion of tumor samples (103 of 261, 39%) showed genomic loss in at least one of the complex associated genes (Figure 2A). KEAP1 was the most frequently disrupted complex component, undergoing genomic loss in 23% of lung tumors analyzed (Figure 2A). Strikingly, 69% (71 of 103) of the tumors harboring copy number alterations had alterations affecting only one of the genes assessed. Gene expression analysis revealed aberrant expression in at least one of the complex component genes in 40% of tumors analyzed (19 of 48), and of those with aberrant expression 84% (16 of 19) had only one of the genes affected (Figure 2A). At the expression level, RBX1 was the most frequently altered complex component, having under expression in 21% of tumors analyzed (Figure 2A). We also detected a high frequency of IKBKB DNA copy number gain and mRNA overexpression (23% and 35% of tumors, respectively) (Figure 2A). When IKBKB status was taken into account, the frequency of NSCLC disruption at any of the KEAP1, CUL3, RBX1 and/or IKBKB loci rose to 54% (141 of 261 tumors, Figure 2A) at the gene copy number level and 63% (30 of 48 tumors, Figure 2A) at the expression level. Even with the inclusion of IKBKB, the majority of tumors exhibited complex or IKBKB genetic disruption at only one gene locus (Figure 2B and 2C). Frequent genetic disruption of E3-ubiquitin ligase complex components and IKBKB (73%) was also evident in NSCLC cell lines (see Table, Supplemental Digital Content 4, which describes complex disruption in NSCLC cell lines).
Figure 2.
Frequent disruption of the KEAP1 E3-ligase complex components and IKBKB in NSCLC. (A) Summary of DNA copy number and gene expression alterations in NSCLC tumors. (B) Copy number analysis of 261 lung tumors revealed frequent loss of KEAP1, RBX1, and CUL3, as well as frequent gain of IKBKB. Vertical columns indicate individual tumor samples and only samples with ≥1 alterations are shown. (C) mRNA expression profiles for 48 lung tumors revealed frequent underexpression of complex components and overexpression of IKBKB. Expression was considered altered if tumor/matched non-malignant tissue was changed >2 fold. (D) KEAP1, CUL3, and IKBKB exhibit statistically significant differences in copy number alteration patterns between AC and SCC (Fisher's exact test, p < 0.01).
To determine whether complex component and IKBKB gene dosage alterations are regulating gene expression, we integrated DNA copy number and gene expression data for the complex genes, and IKBKB in an additional set of 49 NSCLC tumors. KEAP1, RBX1 and CUL3, expression was significantly lower in tumors with genomic loss compared to those without loss (U test, p=0.00076, p=0.00116, and p=0.00339, respectively), whereas IKBKB gene expression was elevated in tumors with gain compared to those without (U test, p=0.0143) (see Figure, Supplemental Digital Content 5). These findings demonstrate that dosage alterations affect mRNA expression levels, and therefore, likely contribute to E3-ligase complex disruption.
In addition to the tumor data generated using array comparative genomic hybridization, we analyzed copy number profiles derived from publically available SNP array data from the dbGaP Genotypes and Phenotypes database for an additional 383 pairs of NSCLCs and matched non-malignant tissue. Consistent with our findings, frequent genomic disruption to the E3-ubiquitin ligase complex components and IKBKB was observed in this external cohort, (see Table, Supplemental Digital Content 4, which describes complex disruption in dbGAP tumors). 34% of the additional 383 tumors harbored DNA alterations encompassing at least one complex component or IKBKB. Furthermore, we interrogated the copy number status of KEAP1, CUL3, RBX1, and IKBKB genes in the Broad Institute's Tumorscape database 30. This revealed that CUL3 was significantly deleted in 12% of all 3131 tumors in the database and in 13% percent of all NSCLC specimens (n=733), of which 5.4% had focal CUL3 deletions. Similarly, IKBKB was significantly amplified in 22% of the all tumors and 28% of NSCLC specimens, of which 12% contained focal IKBKB DNA amplifications. KEAP1 and RBX1 were not significantly deleted in the Tumorscape database.
Differences in gene disruption pattern in squamous and adenocarcinoma subtypes
As distinct patterns of DNA alterations exist for adenocarcinoma (AC) and squamous cell carcinoma (SCC), we sought to determine whether complex component disruption displays subtype specific patterns of alteration. While both subtypes of NSCLC showed high frequency of complex component and IKBKB gene disruption, the pattern of gene disruption differed between these subtypes (Figure 2D and see Table, Supplemental Digital Content 6, which describes complex disruption in AC and SCC tumors). KEAP1 loss appears to be the main mechanism of complex disruption in lung AC, accounting for 64% of the cases that showed disruption (see Table, Supplemental Digital Content 6) and is more prevalent in AC than in the SCC subtype (Fisher's exact test, p = 0.008), in which it is lost in only 16% of cases with component disruption (see Table, Supplemental Digital Content 6). In contrast, CUL3 loss and IKBKB gain occurred more often in the SCC than AC subtype of lung tumors (Fisher's exact test, p = 4.926×10−6 and p = 8.446 × 10−7, respectively; see Table, Supplemental Digital Content 6). The increase of IKBKB gene copy number occurred in 57% of the disrupted SCC cases and only 26% of disrupted AC cases.
Functional consequences of genetic complex disruption
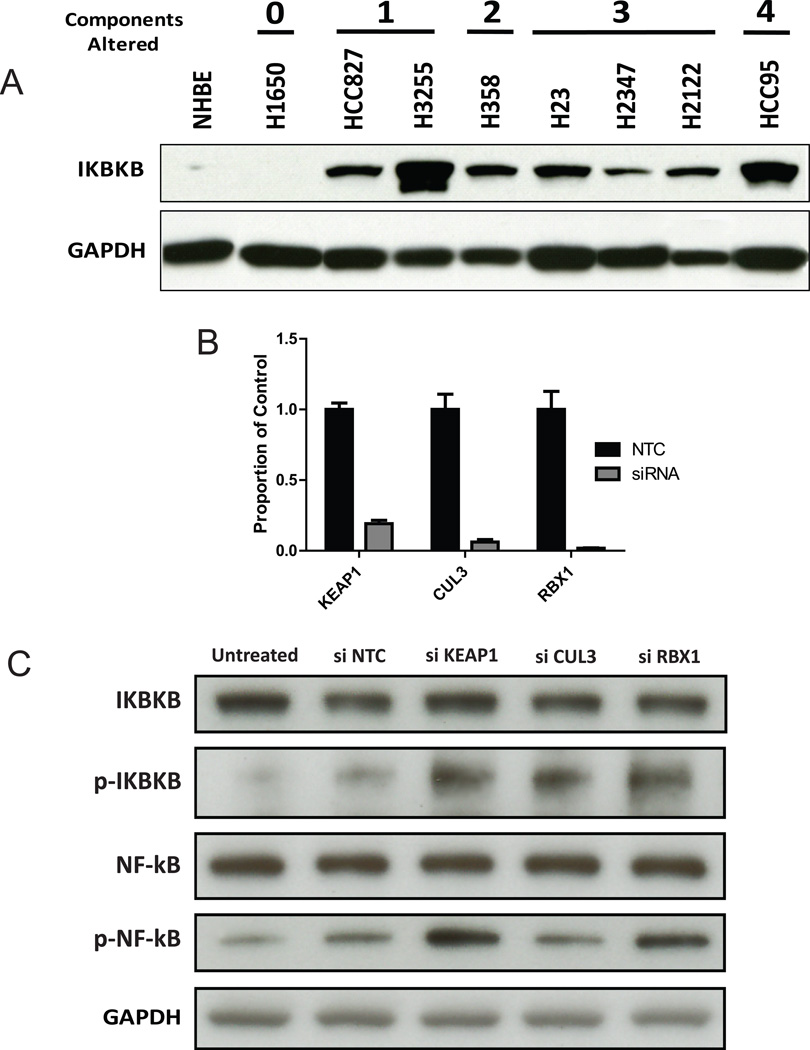
Since IKBKB protein levels would be directly affected by E3-complex function, we assessed IKBKB protein levels in NSCLC cell lines with and without complex disruption to measure the consequence of complex disruption. Immunoblotting for IKBKB in non-malignant human bronchial epithelial (NHBE) cells and a panel of 8 NSCLC cell lines revealed elevated expression levels in lines with genomic loss of KEAP1, CUL3, RBX1, or gain of IKBKB (Figure 3A). To directly assess the effect of complex component integrity on IKBKB and NF-κB activity, we performed siRNA mediated knockdowns of KEAP1, CUL3 and RBX1 in non-malignant, bronchial epithelial cells. We achieved at least 80% knockdown for all three genes and observed an increase in phospho-IKBKB and phospho-NF-κB levels in the knockdowns relative to the NTC, providing evidence that complex disruption directly regulates NF-κB signaling (Figures 3B and 3C).
Figure 3.
IKBKB protein expression in NSCLC. (A) Western blot depicting IKBKB protein expression in eight cell lines with varying degrees of genetic disruption to KEAP1 E3-ligase complex components and/or IKBKB (as determined from copy number profiles of 90 NSCLC cell lines). NHBE is a non-malignant human bronchial epithelial lung line that provides a baseline for IKBKB expression. The number of disrupted genes for each line is indicated above the cell line. (B) qPCR assessment of mRNA expression levels for KEAP1, RBX1 and CUL3 after siRNA-mediated knockdown relative to a non-targeting control (siNTC). Error bars represent the standard error in relative expression across all biological replicates. (C) Western blot depicting the effects of transient siRNA knockdown of KEAP1, RBX1, and CUL3, on total and phospho- IKBKB and NF-κB protein levels.
Staining for IKBKB in NSCLC tumors also revealed protein expression in both complex disrupted and undisrupted tumors (Supplemental Digital Content 7); however, the vast inter- and intratumor heterogeneity in staining limited our ability to identify any significant correlations between IKBKB protein levels and E3-complex genetic disruption.
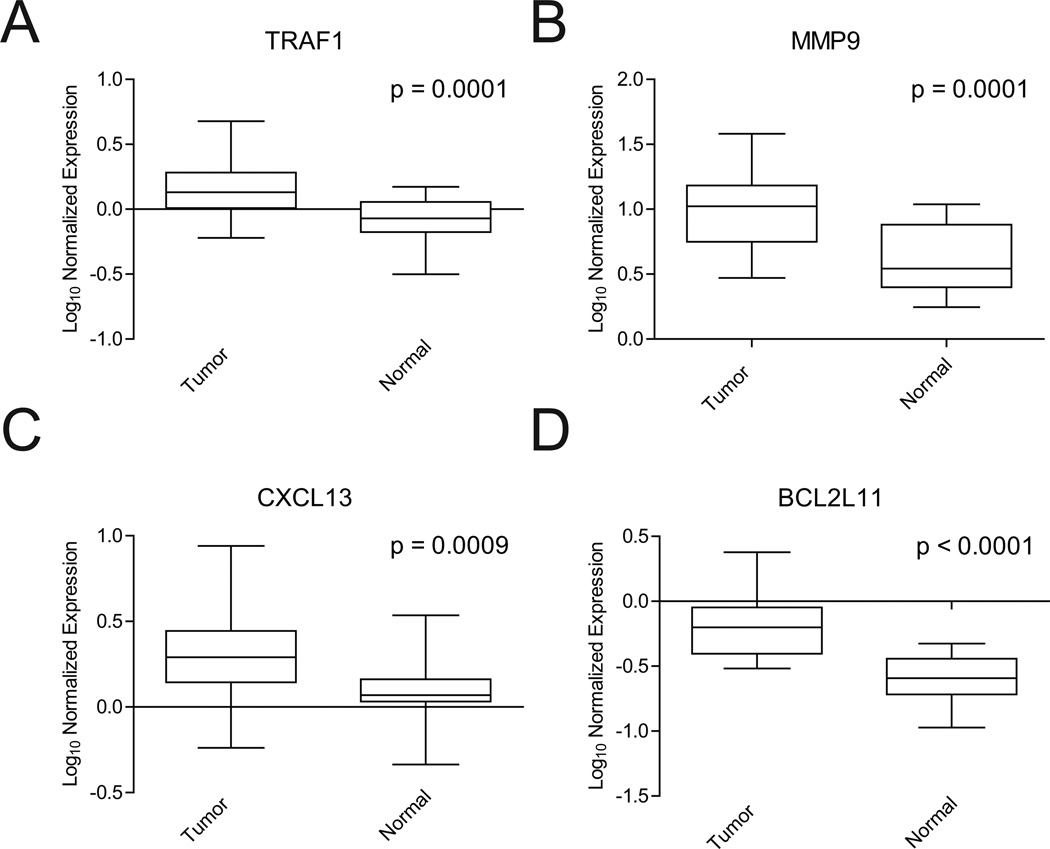
To determine whether E3-ligase complex component underexpression activates NF-κB signaling in lung tumors, expression of nine well characterized transcriptional targets of NF-κB were analyzed. Of the nine genes, five - BCL2L11, CXCL13, CXCR4, MMP9, and TRAF1 - were significantly up-regulated in tumors with underexpression of a complex component or overexpression of IKBKB as compared to their matched non-malignant lung tissue (Figure 4). Similarly, assessment of the same NF-κB target genes following siRNA confirmed these genes to be overexpressed relative to non-targeting controls (data not shown).
Figure 4.
Activation of NF-κB targets in tumors with aberrant expression of KEAP1 E3-ligase complex components and/or IKBKB. (A–D) Box and whisker plots demonstrating four examples of NF-κB target genes (BCL2L11, CXCL13, MMP9, TRAF1) that are significantly upregulated in tumors with underexpression of KEAP1, RBX1, and/or CUL3 or overexpression of IKBKB, relative to matched non-malignant lung tissues (Wilcoxon sign rank test, p ≤ 0.001). Copy number status of the NF-κB target gene loci was determine in these tumors to ensure that overexpression was not due to dosage changes of the target genes.
Pharmacological inhibition of IKBKB
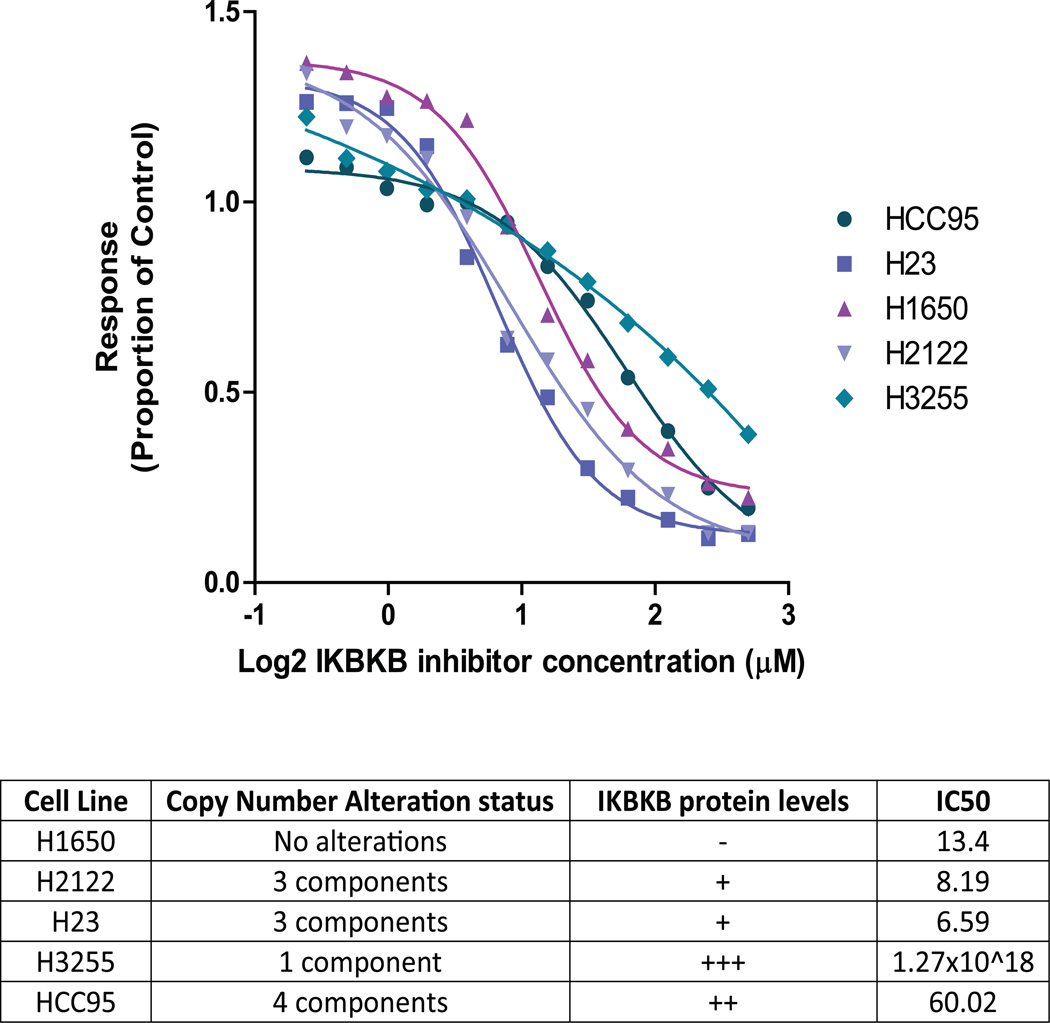
Overexpression of IKBKB can contribute to a malignant phenotype through activation of the NF-κB signaling pathway and consequential effects on cell growth. As such, we hypothesized cells dependent on this pathway would be more sensitive to IKBKB inhibition than those without complex alterations and normal IKBKB protein levels. IKBKB inhibition experiments revealed H1650, an AC line without genomic loss of any complex component, had reduced sensitivity to IKBKB inhibition (IC50 13.41) than H2122 and H23 (IC50 8.19 and 6.59, respectively) which have multiple components altered (student's t-test, p < 0.05) (Figure 5). H3255 and HCC95 showed greater insensitivity to inhibition than H1650 (student's t-test, p < 0.05), however these two cell lines have the highest IKBKB protein expression levels of the cell lines we tested, which likely contributed to their relative resistance to inhibition. Replicate experiments were highly reproducible, with similar trends in sensitivity observed across all replicates.
Figure 5.
Pharmacological inhibition of IKBKB in NSCLC cells. Alamar Blue cell viability assays were performed to measure the effect of IKBKB inhibition by a cell permeable, competitive ATP inhibitor, IKK-2 inhibitor IV on 5 NSCLC cell lines. H2122, H23, H3255, and HCC95 harboured genetic alterations to either IKBKB and/or one or more of the KEAP1 E3-ligase complex components while H1650 was not altered at the DNA level.
Discussion
The NF-κB pathway is aberrantly activated in the majority of lung cancers and is essential in mouse models of lung tumorigenesis 10–13. NF-κB signaling contributes to tumorigenesis via its promotion of cell proliferation and survival 7–9. In order for NF-κB to become active, inhibition by IκB must be released. This is achieved through phosphorylation of IκB by the kinase, IKBKB 14; hence, IKBKB has a critical role in NF-κB activation 6, 14. In fact, constitutive IKBKB activity has been postulated to drive the aberrant NF-κB activation observed in cancer 8. Despite what is known about the cascade of protein signaling events that result in NF-κB activation, the genetic mechanisms responsible for the aberrant activation of NF-κB signaling in lung cancer are not well understood. In this study, we hypothesized that genetic disruption and loss of function of the KEAP1 E3-ubiquitin ligase complex, which regulates IKBKB protein levels, is a major mechanism of IKBKB accumulation and consequential NF-κB activation in lung cancer.
Our results provide evidence that somatic E3-ligase complex disruption is a prominent genetic mechanism of NF-κB activation in lung cancer that compromises the ability of cells to degrade the NF-κB activator, IKBKB. We discovered a remarkably high frequency of both genetic disruption and gene expression changes for the genes encoding E3-ligase protein components (KEAP1, RBX1, and CUL3) as well as the gene encoding the complex's oncogenic substrate, IKBKB. We found that genetic disruption of the complex genes alters mRNA expression and results in elevated IKBKB protein levels, demonstrating the consequence of complex disruption. Moreover, we demonstrated evidence of NF-κB activation in complex compromised lung tumors and showed the importance of IKBKB protein expression in driving the lung cancer phenotype.
Although genetic and epigenetic disruption of KEAP1 has been reported in lung cancer before, to our knowledge, this is the first study to comprehensively characterize somatic gene dosage alterations to the CUL3, RBX1, and IKBKB loci in a large cohort of clinical lung tumors. The strikingly high frequency of copy number and gene expression alterations observed in our study highlights the importance of these E3-ubiquitin ligase complex components and also IKBKB in lung cancer. The recurrent nature of DNA copy number alterations at the complex component loci and their effects on gene expression are strong evidence that these genes are targeted for dosage alterations as opposed to passengers of alterations targeting other genes. In addition, the high proportion of disrupted lung tumors observed to have genetic alterations affecting a single component only, at both the copy number (67%, Figure 2B) and gene expression levels (73%, Figure 2C), suggests that disruption of only a single complex component is sufficient to compromise complex function and promote NF-κB signaling through abnormal IKBKB accumulation. Interestingly, we observed differential complex component disruption patterns in AC and SCC subtypes (Figure 2D). Although E3-ubiquitin ligase complex disruption occurs in both subtypes, the differences in the component genes preferentially altered suggests that complex disruption is achieved by different means.
Examination of IKBKB protein levels in NSCLC cell lines revealed high expression in lines harboring genetic disruption to at least one complex component or IKBKB, whereas the non-malignant lung line (NHBE) and a line without genetic disruption (H1650) showed very low or undetectable levels. The E3-ligase complex was considered to be genetically intact in H1650 as neither underexpression of KEAP1, RBX1, and CUL3, or overexpression of IKBKB relative to NHBE cells was observed (data not shown). This suggests there are no genetic or epigenetic alterations affecting the complex components or IKBKB in this cell line and the observed IKBKB levels were consistent with H1650 having a functioning E3-ligase complex, supporting our hypothesis. Therefore, in addition to affecting gene expression, copy number losses of the loci coding for complex components and gains of IKBKB appear to influence IKBKB protein expression. A trend towards higher IKBKB expression in lines with more complex components/IKBKB alterations was not evident, suggesting genetic disruption of a single component is sufficient to result in loss of complex function and IKBKB accumulation (Figure 3A). This finding is consistent with the observation that the majority of tumors exhibiting complex disruption have only one complex component altered, further supporting the idea that single component disruption is sufficient to produce an oncogenic effect.
Given our hypothesis, we focused on measuring gene dosage alterations that could account for disruption of the E3-ubiquitin ligase complex and its downstream consequences. However, it is conceivable that other genetic and epigenetic mechanisms could also contribute to silencing of the E3-ubiquitin ligase complex components thereby contributing to the observed accumulation of IKBKB and aberrant NF-κB signaling. Mutations and hypermethylation of KEAP1 have been described and these events can result in downregulation of KEAP1 expression 15, 17, 18, 21. The COSMIC (Catologue of Somatic Mutations in Cancer) database has compiled mutation status for thousands of genes in cancer genomes 31. As KEAP1 mutations are known to exist in lung cancer (albeit at low frequencies), we searched COSMIC for reported mutations in RBX1, CUL3 and IKBKB. No RBX1 mutations have been reported in over 170 cancer samples analyzed, while CUL3 and IKBKB mutations have been identified in three of 173 and 5 of 660 cancer samples, respectively, of which only one IKBKB mutation was in lung cancer. Thus, the rarity of mutations in these genes in lung cancer specifically, suggests they are unlikely to be mutated and unlikely to play a role in complex disruption in the lung cancer cell lines we assessed. Furthermore, we have shown that DNA alterations at the KEAP1, CUL3, RBX1, and IKBKB loci are associated with concurrent expression changes, providing evidence that gene dosage alterations are a significant mechanism driving complex disruption at the genetic level.
We have conclusively demonstrated elevated IKBKB protein expression in NSCLC cell lines with complex disruption, however, measuring this effect directly in tumor tissue sections was not a straightforward task due to the extent of heterogeneity in tumor staining intensity across and within individual tumors. The variation in tissue distribution of stained cells is exemplified by two tumor samples shown in Supplemental Digital Content 7B (genetic complex disruption) and 7C (no genetic complex disruption), and likely reflects the heterogeneous nature of lung tumor specimens. Due to this innate tumor heterogeneity, unlike cell lines, we were unable to conclude whether or not there was a significant correlation between E3-ligase complex disruption and IKBKB protein levels in vivo.
To investigate the direct consequence of complex disruption on IKBKB accumulation and NF-κB activity, we performed siRNA knockdowns on the individual complex coding genes (KEAP1, RBX1, and CUL3). Consistent with our hypothesis, we observed elevated levels of activated IKBKB and NF-κB upon complex component disruption in HBEC cells (Figure 3C). Since phospho-NF-κB is an indicator of active NF-κB signaling, these results clearly illustrate the functional consequence of E3-ligase complex disruption. Our work provides evidence to support the hypothesis that genetic loss of the complex component encoding genes causes downregulation in their expression, and that loss of expression of these genes results in increased levels of activated IKBKB and NF-κB.
A number of reports have detailed the critical role of IKBKB protein in driving NF-κB activation, and the importance of IKBKB to cancer cell viability is emphasized by the development of IKBKB inhibitors as a strategy for tempering NF-κB signaling 8, 14, 22. We found that IKBKB inhibition reduced NSCLC cell viability and that cells without complex or IKBKB disruption, which we hypothesized to be less dependent on IKBKB expression for growth, were indeed more insensitive to IKBKB inhibition, as were cells with high endogenous levels of IKBKB protein (Figure 5). In addition to cell experiments to verify the importance of E3-ligase complex disruption in lung cancer, we analyzed the expression levels of several NF-κB target genes in tumors with complex disruption to measure its effect on NF-κB activity. Despite the possibility that other mechanisms could also contribute to the transcription of the NF-κB target genes, we observed a significant increase in the expression of NF-κB target genes in complex compromised tumors (Figure 4). We also observed elevated expression of NF-κB target genes following knockdown of the E3-ligase complex components. Together, these findings support our hypotheses and demonstrate the biological significance of complex disruption and subsequent IKBKB overexpression in lung cancer biology.
Collectively, our analyses have revealed remarkably frequent genetic disruption and aberrant expression not only of KEAP1, but all members of the KEAP1 E3-ubiquitin ligase complex and IKBKB in lung cancer. We have shown that IKBKB protein expression is elevated in NSCLCs with genetic loss of KEAP1, CUL3 or RBX1 or gain of IKBKB, and that knockdown of complex components leads to an accumulation of active IKBKB and NF-κB, thereby demonstrating the functional consequence and significance of complex disruption. We have also provided evidence of NF-κB activity, a downstream effect of IKBKB accumulation, in complex disrupted tumors and cell lines. Interestingly, it appears that AC and SCCs of the lung acquire copy number alterations to different components of the E3-complex or IKBKB which suggests the genetic mechanisms of complex disruption that promote NF-κB activation may be subtype specific. Our findings suggest that prominent genetic disruption to the E3-ubiquitin ligase complex and its oncogenic substrate, IKBKB, play a major role in driving the aberrant NF-κB activation that is characteristic of lung tumorigenesis.
Supplementary Material
Acknowledgements
The authors would like to thank Chad Malloff, Greg Stewart, and Ewan Gibb for insightful comments regarding the manuscript.
Support
This work was supported by funds from the Canadian Institutes for Health Research (CIHR; MOP 86731, MOP 94867), Canadian Cancer Society (CCS20485 and CCS20527), NCI Early Detection Research Network (EDRN; 5U01 CA84971-10), and the Canary Foundation. KLT, LAP, RC, IMW and WWL are supported by scholarships from the CIHR, the Michael Smith Foundation for Health Research, Vanier Canada Graduate Scholarship, and the University of British Columbia Interdisciplinary Oncology Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Digital Content
Supplemental Digital Content 1. doc – Tumor demographics
Supplemental Digital Content 2. doc – NSCLC cell lines analyzed
Supplemental Digital Content 3. doc – NF-kB target genes
Supplemental Digital Content 4. doc – Cell line and dbGaP disruption frequencies
Supplemental Digital Content 5. doc – Correlation between copy number and expression in clinical lung tumors
Supplemental Digital Content 6. doc – Differential disruption in AC vs SCC tumors
Supplemental Digital Content 7. doc – IHC in clinical tumors
References
- 1.Cullinan SB, Gordan JD, Jin J, et al. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi A, Kang MI, Okawa H, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Blake DJ, Singh A, Kombairaju P, et al. Deletion of Keap1 in the lung attenuates acute cigarette smoke-induced oxidative stress and inflammation. Am J Respir Cell Mol Biol. 2010;42:524–536. doi: 10.1165/rcmb.2009-0054OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee DF, Kuo HP, Liu M, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karin M, Cao Y, Greten FR, et al. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 8.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y, Bai L, Chen W, et al. The NF-kappaB activation pathways, emerging molecular targets for cancer prevention and therapy. Expert Opin Ther Targets. 2010;14:45–55. doi: 10.1517/14728220903431069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto T, Sanda T, Asamitsu K. NF-kappa B signaling and carcinogenesis. Curr Pharm Des. 2007;13:447–462. doi: 10.2174/138161207780162944. [DOI] [PubMed] [Google Scholar]
- 11.Tang X, Liu D, Shishodia S, et al. Nuclear factor-kappaB (NF-kappaB) is frequently expressed in lung cancer and preneoplastic lesions. Cancer. 2006;107:2637–2646. doi: 10.1002/cncr.22315. [DOI] [PubMed] [Google Scholar]
- 12.Meylan E, Dooley AL, Feldser DM, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basseres DS, Ebbs A, Levantini E, et al. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010;70:3537–3546. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid JA, Birbach A. IkappaB kinase beta (IKKbeta/IKK2/IKBKB)--a key molecule in signaling to the transcription factor NF-kappaB. Cytokine Growth Factor Rev. 2008;19:157–165. doi: 10.1016/j.cytogfr.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Solis LM, Behrens C, Dong W, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohta T, Iijima K, Miyamoto M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 17.Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, An J, Ji F, et al. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Kohno T, Otsuka A, Girard L, et al. A catalog of genes homozygously deleted in human lung cancer and the candidacy of PTPRD as a tumor suppressor gene. Genes Chromosomes Cancer. 2010 doi: 10.1002/gcc.20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki T, Maher J, Yamamoto M. Select Heterozygous Keap1 Mutations Have a Dominant-negative Effect on Wild-type Keap1 In Vivo. Cancer Research. 2010 doi: 10.1158/0008-5472.CAN-10-2939. epub. [DOI] [PubMed] [Google Scholar]
- 21.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 22.Bivona TG, Hieronymus H, Parker J, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garnis C, Lockwood WW, Vucic E, et al. High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- 24.Gazdar AF, Girard L, Lockwood WW, et al. Lung cancer cell lines as tools for biomedical discovery and research. J National Cancer Institute. 2010 doi: 10.1093/jnci/djq279. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockwood WW, Chari R, Coe BP, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med. 2010;7:e1000315. doi: 10.1371/journal.pmed.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 27.Wong KK, deLeeuw RJ, Dosanjh NS, et al. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockwood WW, Chari R, Coe BP, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 30.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.