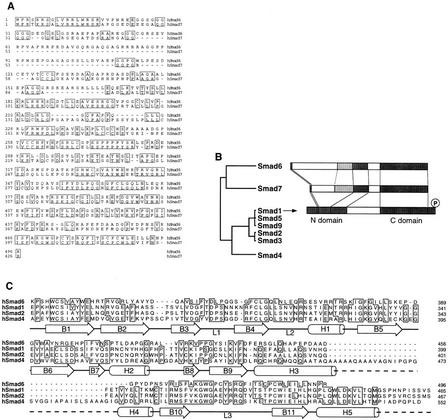
Figure 1.
Structure of Smad6. (A) Alignment of the predicted amino acid sequence of human Smad6 and Smad7. Identical residues are boxed. (B) Smad homology tree and schematic comparison of the structures of Smad6, Smad7, and Smad1. The amino and carboxy-terminal homologous regions (N- and C-domain) are darkly shaded. A region conserved only between Smad6 and Smad7 is lightly shaded. (C) Alignment of C-domain sequences of Smad6 and other Smads, indicating the secondary structure elements of the Smad4 C-domain (Shi et al. 1997). In the crystal structure of Smad4 this region forms an α-helix (α-helix 5) that associates with α-helices 3 and 4 in a three-helix bundle. This structure contributes the formation of a Smad4 homotrimer by interacting with loops 1 and 2 of the adjacent monomer. The region corresponding to loops 1 and 2 in Smad6 is also very divergent from Smad4 and the receptor-regulated Smads. However, most of the components that constitute the β-sandwich core structure of the Smad4 C-domain are conserved in Smad6.