FIGURE 6:
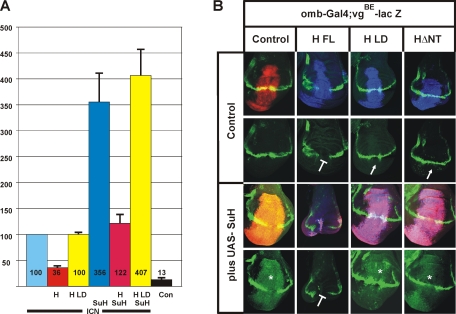
Mutant Hairless fails to assemble functional repressor complexes. (A) Effects of wild-type or mutant Hairless protein, in the absence or presence of Su(H), on Notch ICN-mediated expression from the Notch reporter (NRE) were studied in Drosophila S2 cell culture. S2 cells were transiently transfected with constructs indicated; empty vector was used as a negative control. Luciferase activity is represented on the y-axis, and transfection efficiency was normalized by cotransfection of the Renilla plasmid. Values for NRE expression in the presence of Notch ICN were normalized to 100% (lane 1). (B) Effects of the overexpression of wild-type and mutant Hairless constructs, singly or together with Su(H), on in vivo expression of the Notch target gene vestigial. Wild-type and mutant UAS constructs as indicated were induced in the central region of wing imaginal disks using the omb-Gal4 driver line and visualized by antibody staining. UAS-GFP served as control (red). Expression of vgBE-lacZ reporter is expected along the dorsoventral boundary and shown in green (bottom). In the merged picture (top), overlap appears yellow. Overexpressed Su(H) is shown in red, and Hairless protein is shown in blue; overlap appears magenta, and with vgBE-lacZ (green) it appears white. Overexpression of Su(H) causes an overgrowth of the disk and an activation of vgBE-lacZ (asterisk). Conversely, overexpression of HFL results in a repression of vgBE-lacZ (blunt arrow). However, overexpression of the Hairless mutant constructs HΔNT or HLD has little effect on vgBE-lacZ expression, reflecting the loss of Su(H)-binding activity (arrow). A combination of HFL with Su(H) strongly affects growth of the wing disk and vgBE-lacZ expression. In contrast, a combination of Su(H) with either HΔNT or HLD results in overgrowth and vgBE-lacZ induction that resembles the sole Su(H) overexpression, indicating that these Hairless mutants fail to form repressor complexes in vivo.