Figure 3.
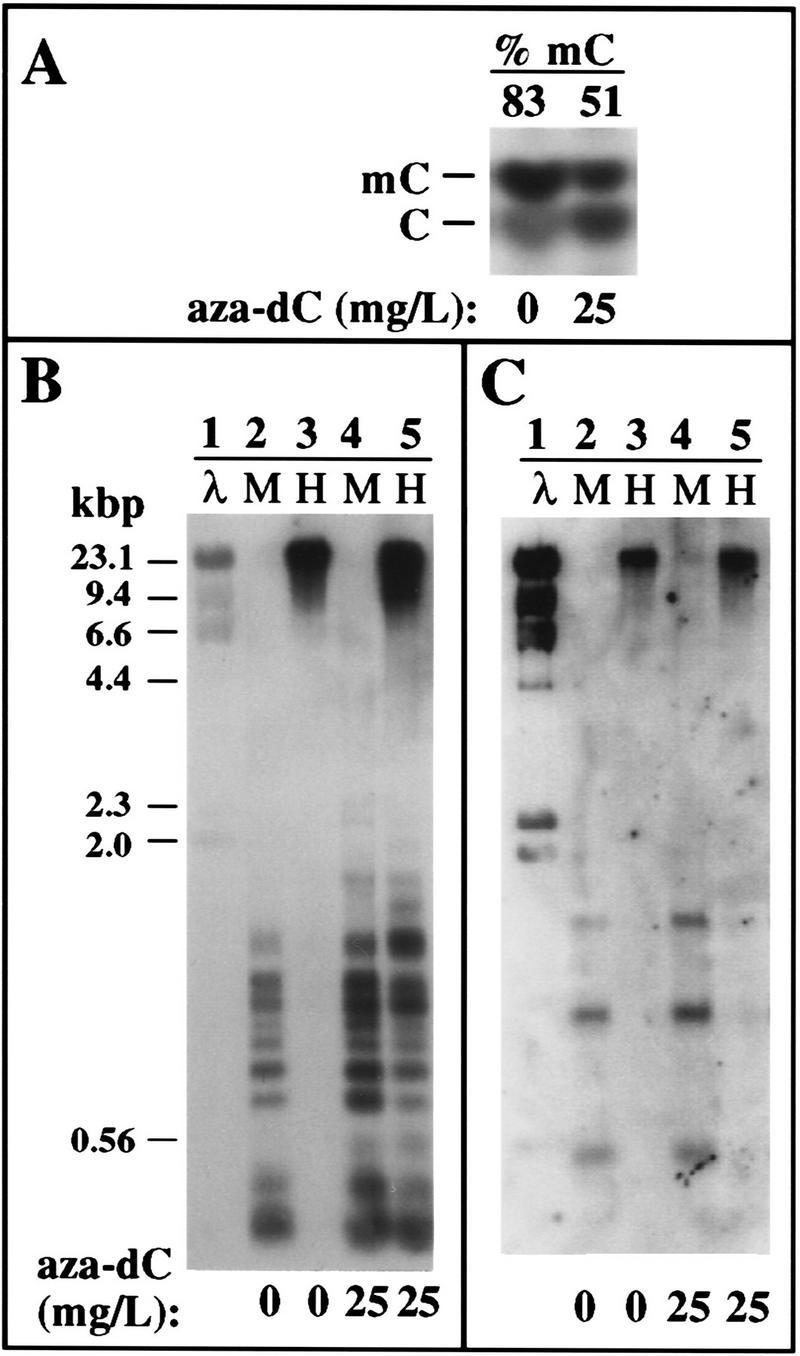
Effects of aza-dC on global and rRNA gene methylation. (A) Aza-dC treatment reduces genomic methyldeoxycytidine levels ∼30%. Genomic DNA isolated from control or aza-dC treated B. napus plants was cleaved to completion with TaqI. TaqI is insensitive to cytosine methylation and cleaves at the recognition sequence TCGA to leave a 5′ overhanging cytosine that can be end-labeled with T4 kinase and γ-labeled 32P[ATP]. Following complete DNase digestion, samples were subjected to TLC and autoradiography. Numbers at the top indicate the amount of methyldeoxycytidine (% of total deoxycytidine) in control and treated plants based on quantitation by phosphorimaging. (B,C) Aza-dC treatment reduces cytosine methylation in the vicinity of the rRNA gene promoter. DNA from control and aza-dC-treated B. napus plants was digested with the isoschizomers MspI (M) or HpaII (H), then subjected to electrophoresis, Southern blotting, and hybridization to a probe spanning −166 (relative to the transcription start site, +1) to the beginning of the 18S rRNA coding region (B). Aza-dC caused increased susceptibility to the methylation-sensitive enzyme Hpa II (cf. lanes 3 and 5). (C) Same blot was stripped and reprobed with a labeled 5S RNA gene fragment to verify that similar amounts of DNA were loaded in all lanes. HindIII-digested λ DNA markers were run in lane 1 and are visible on the autoradiogram because a small amount of λ DNA was mixed with the rDNA probe fragments in the labeling reactions.
