Figure 5.
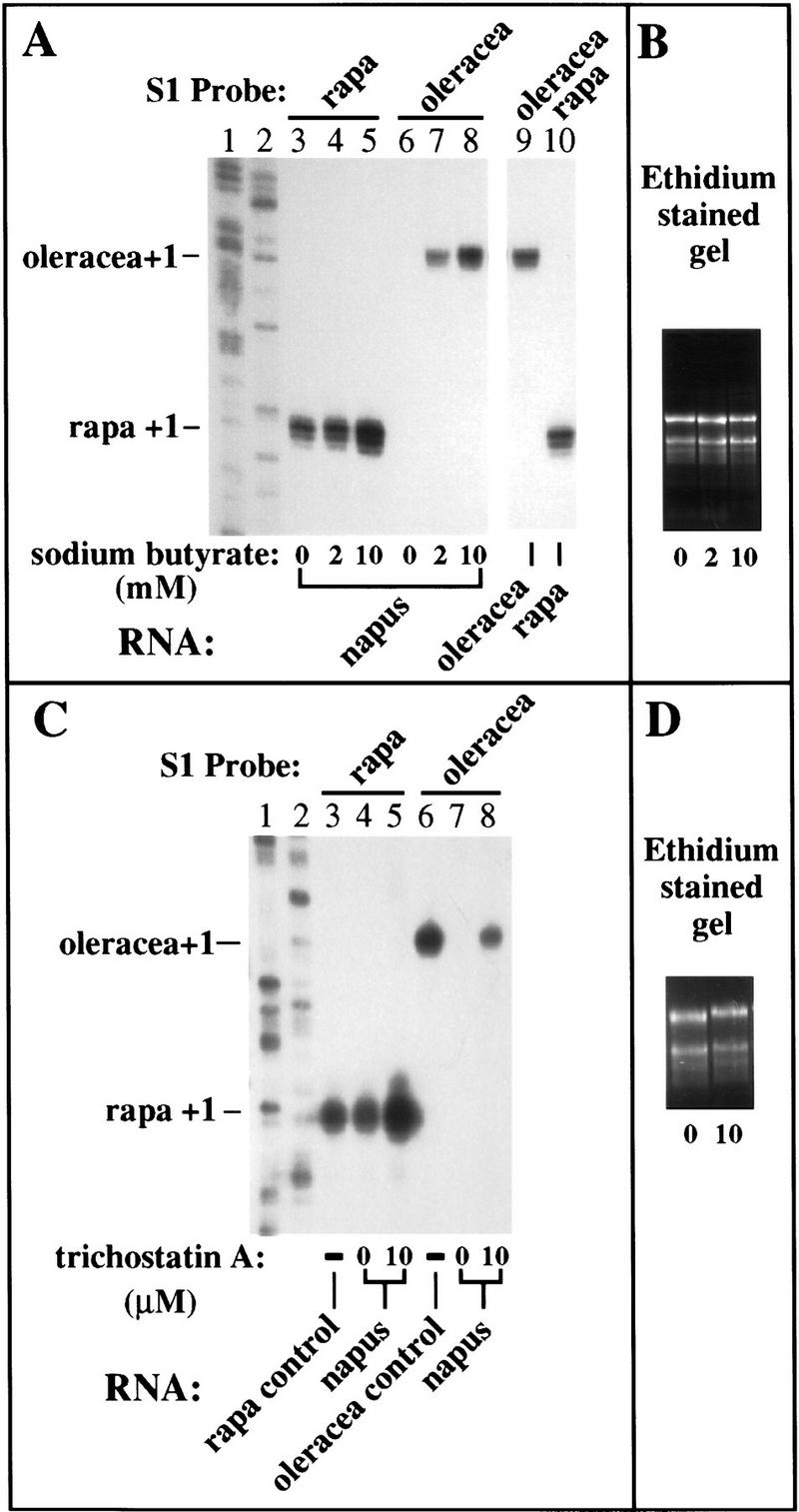
Inhibitors of histone deacetylation derepress silenced rRNA genes. (A,B) B. napus seeds were germinated on medium containing 0, 2, or 10 mm sodium butyrate and total RNA (10 μg) from two-week-old plants was subjected to S1 nuclease protection using the B. rapa (lanes 3–5,10) and B. oleracea-specific (lanes 6–9) probes. S1 protected products using RNA from the diploids B. oleracea and B. rapa were run on the same gel as controls to show that the two probes had equivalent specific activity (lanes 9,10). (Lanes 1,2) Sequencing ladders used as size markers. Note that the normally silent B. oleracea rRNA genes were activated by sodium butyrate treatment (cf. lanes 6 and 8) and dominant B. rapa rRNA gene transcript levels were also up-regulated two- to threefold (cf. lanes 3 and 5). (B) An ethidium bromide-stained agarose gel of RNA aliquots from plants subjected to the various treatments to verify that equivalent amounts of RNA were probed to yield the results in A. (C,D) In an experiment similar to that shown in A, B. napus seeds were germinated on medium containing 0 or 10 μm trichostatin A and RNA from two-week-old plants was subjected to S1 nuclease protection analysis using B. rapa and B. oleracea-specific probes. RNA from the diploid progenitors served as controls in lanes 3 and 6. Sequencing ladders served as size markers in lanes 1 and 2. Like sodium butyrate and aza-dC, trichostatin A also derepressed the normally silent B. oleracea genes (cf. lanes 7 and 8) and up-regulated B. rapa rRNA transcript levels (cf. lanes 4 and 5). (D) An ethidium-stained gel to confirm that equivalent amounts of RNA were tested to yield the results shown in C.
