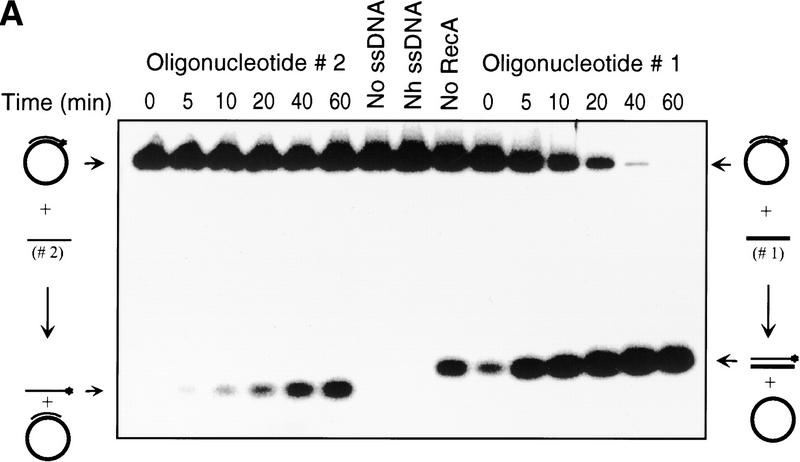
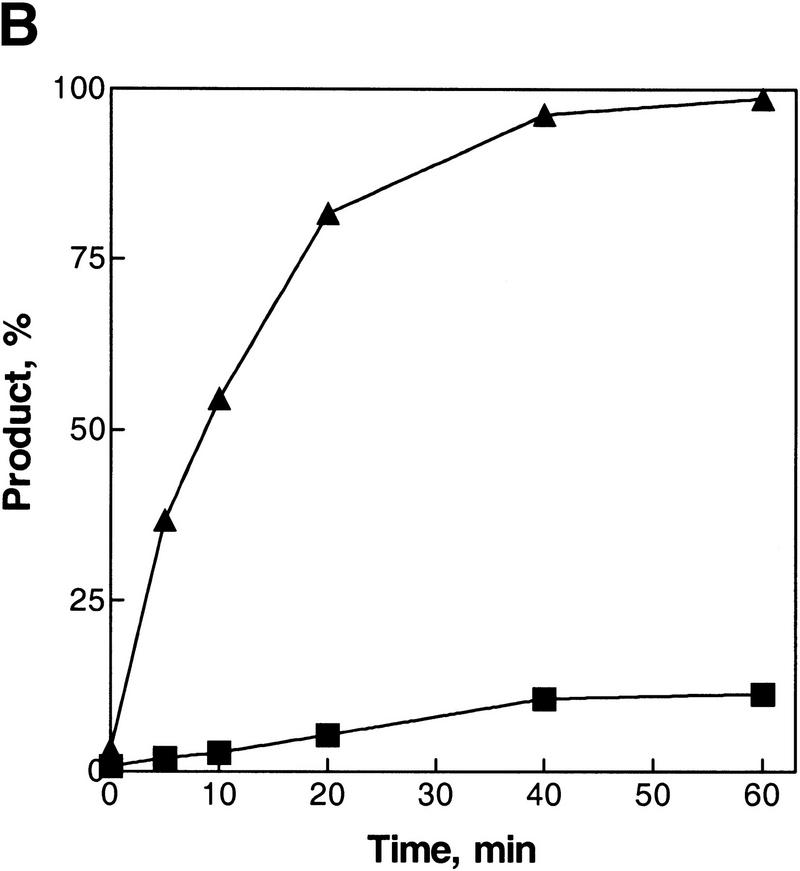
Figure 2.
Asymmetry of inverse DNA strand exchange. (A) Time courses for the inverse reactions depicted in Fig. 1. The reactions were carried out as described under Materials and Methods. Concentrations of oligonucleotides 1 and 2 were 10-fold higher than the molar (molecule) concentration of the M13 ssDNA–oligonucleotide 2 duplex substrate. The lanes marked No RecA and No ssDNA designate control reactions performed without either RecA protein or oligonucleotide 1, respectively. The lane marked Nh ssDNA designates the control reaction using nonhomologous ssDNA (63-mer oligonucleotide 3) instead of the homologous ssDNA. Control reactions were conducted for 60 min at 37°C. To prevent annealing of the potentially displaced oligonucleotide 2 back to the M13 ssDNA, a 10-fold molar (molecular) excess of cold oligonucleotide 2 was added to the control reactions just before deproteinization. (B) Data from A, quantified and plotted (▴) Oligonucleotide 1; (█) oligonucleotide 2.