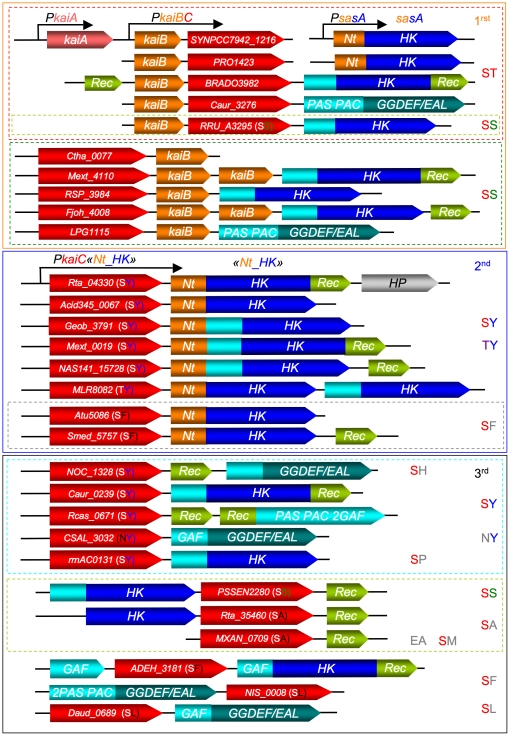
Figure 4. Schematic representation of predicted KaiC genetic organization compared to cyanobacterial KaiABC-SasA «clock system».
(SasA is found isolated in Cyanobacteria genomes). This representation is based on the phylogeny of predicted KaiC according to TULIP tree (Fig. S6). The first clustering corresponds to colour and KaiC-context group name (1rst, 2nd, 3rd) according to the text and exhibits nature of the phosphorylation sites (ST, SS, SY, TY etc…). KaiC proteins (red colour) have been named according to their encoding gene position in database. KaiC neighbouring proteins were represented according to their proteic domain contents: HK, histidine kinase domain constituted of a HisKA and an HATPase_c domains; REC, single domain receiver protein; PAS, PAS domain; PAC, PAC domain; GAF, GAF domain; GGDEF/EAL, GGDEF and EAL domains. For HK, the N-terminal, PAS, PAC or GAF domains have been replaced by blue-light colour (for details see Fig. S6). Nt_HK: HK with a N-terminal «orange» domain exhibiting similarities with cyanobacterial KaiB protein and kaiB-like N-terminal KaiC-interacting sensory HK SasA. Genes are not drawn on scale.