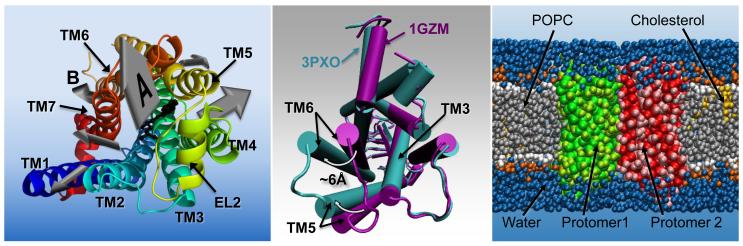
Figure 3.
Three key events in the life of a GPCR. The leftmost panel illustrates ligand recognition, showing the exit pathways of carazolol (black, stick representation) from the crystallographically found binding site in the β2AR (rainbow blue to red by residue number, cartoon representation, viewed from the extracellular side) as determined by the RAMD study [34] reviewed here. The size of the arrow denotes the approximate relative frequency of occurrence of each of the exit pathways during the RAMD. Arrow A is the major exit pathway found by RAMD, and is highlighted as the closest equivalent to the entry pathway observed for NLX into the binding pocket of the DOR, in the cited WTM study [33]. Arrow B is highlighted as the exit pathway found by RAMD that is the closest equivalent to the entry pathway of 2-AG into the cannabinoid receptor, via a cleft between TM6 and TM7, suggested in [32]. The central panel illustrates the largest conformational changes occurring activation using an intracellular view of the aligned structures of inactive rhodopsin (PDBid: 1GZM) [53] and metarhodopsin II (PDBid: 3PXO) [46]. Changes highlighted in the activated structure are the movement of TM6 by ~ 6Å, movement and torsion of TM5. The right-most panel shows a system setup for a coarse-grained simulation of a GPCR homodimer, in an explicit lipid environment. Water is shown in blue, the phosphocholine bilayer is shown in grey (hydrophobic tails) and red/orange (head groups), cholesterol is yellow and the two protomers are shown in red and green.