Abstract
Cells co-express multiple G protein β and γ subunit isoforms, but the extent to which individual subunits associate to form particular βγ complexes is not known. This issue is important because in vivo knockout experiments suggest that specific βγ complexes may have unique functions despite the fact that most complexes exhibit similar properties when assayed in reconstituted systems. This chapter describes how multicolor bimolecular fluorescence complementation (BiFC) can be used in living cells to study the association preferences of β and γ subunits. Multicolor BiFC determines the association preferences of these subunits by quantifying the two fluorescent complexes formed when β or γ subunits fused to amino terminal fragments of yellow fluorescent protein (YFP-N) and cyan fluorescent protein (CFP-N) compete for interaction with limiting amounts of a common γ or β subunit, respectively, fused to a carboxyl terminal fragment of CFP (CFP-C).
One means by which βγ complexes may differ from each other and thereby mediate unique functions in vivo is in the kinetics and patterns of their internalization responses to stimulation of G protein-coupled receptors (GPCRs). Methods are described for imaging and quantifying the internalization of pairs of βγ complexes in response to GPCR stimulation in living cells.
Keywords: multicolor bimolecular fluorescence complementation, heterotrimeric G protein, G protein βγ complex, spectrofluorometer, yellow fluorescent protein, cyan fluorescent protein, fluorescence microscopy, live cell imaging, subcellular targeting, G protein-coupled receptor
1. Introduction
Although most combinations of the 5 G protein β subunits and 12 γ subunits known to be expressed in mammals form dimers with similar abilities to modulate the activities of effector proteins in vitro, specific αβγ combinations appear to be preferred for particular GPCR-G protein signaling pathways in vivo (1). For instance, ribozyme-mediated depletion of γ7 in HEK-293 cells leads to the selective loss of β1 and results in decreased activation of adenylyl cyclase in response to stimulation of β-adrenergic receptors (2, 3). Mice lacking γ7 exhibit increased startle responses and specific decreases in the levels of αolf in the striatum (4). In addition, mice lacking γ3, which are lean and display an increased susceptibility to seizures, display selective decreases in αi3 and β2 (5). In most cases the αβγ heterotrimers that mediate GPCR signaling pathways and the βγ combinations that predominate in particular cell types are not known. The relative amounts of the βγ complexes formed in a cell will depend on the expression levels of the β and γ subunits and on their accessibilities to and relative affinities for each other. Multicolor BiFC enables quantification of the association preferences of β and γ subunits in intact cells.
Multicolor BiFC consists of the simultaneous visualization of the two fluorescent complexes formed when proteins fused to amino terminal fragments of YFP and CFP (YFP-N and CFP-N, respectively) interact with a common binding partner fused to a carboxyl terminal fragment of CFP (CFP-C). The amino terminal fragment of the fluorescent protein contains the chromophore and determines the spectral properties of the complex (6). Therefore, complexes of YFP-N and CFP-C fusion proteins are yellow, whereas those consisting of CFP-N and CFP-C fusion proteins are cyan (See Figure 1). In the methods described here the fluorescent proteins are split at residue 158 such that the amino terminal fragment consists of residues 1-158 and the carboxyl terminal fragment consists of residues 159-238. For competition analysis, we use Cerulean, a modified version of ECFP that is 2.5-fold brighter than ECFP (7), to produce Cer-N fusion proteins, because Cer-N fusions compete more effectively with YFP-N fusions than do CFP-N fusions.
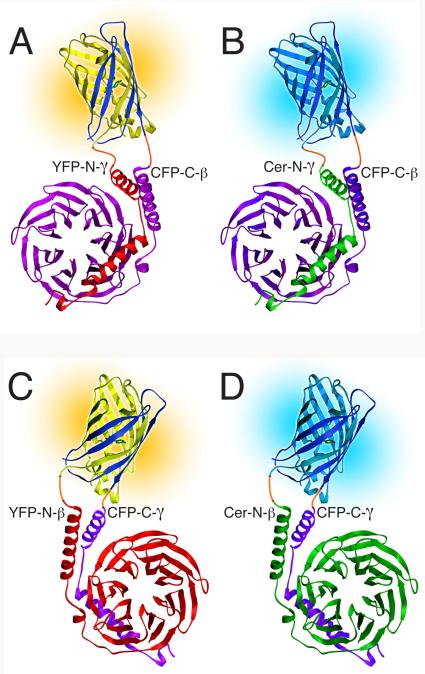
FIG. 1.
Models of fluorescent βγ complexes produced with multicolor BiFC. The split fluorescent protein at the top of each model is joined by linkers (orange) to the βγ dimer at the bottom. The CFP-C fragment (dark blue) is combined with either the YFP-N fragment (yellow) to produce yellow fluorescence or the Cer-N fragment (cyan) to produce cyan fluorescence. (A and B) YFP-N-γ and Cer-N-γ compete for CFP-C-β. β is magenta and γ is red (A) or green (B). (C and D) YFP-N-β and Cer-N-β compete for CFP-C-γ. γ is magenta and β is red (C) or green (D). The structures of the fluorescent protein fragments are adapted from the structure of GFP (16). The structures of β and γ are from the structure of an αt/αi1 chimera complexed with βtγt (17). (Reprinted from (18) with permission from Elsevier.)
To compare the abilities of different γ subunits to compete for the same β subunit, one of the γ subunits (red in Fig. 1A) is fused to the carboxyl terminus of YFP-N (yellow in Fig. 1A) and each of the γ subunits (green in Fig. 1B) is fused to the carboxyl terminus of Cer-N (cyan in Fig. 1B). The β subunit that is competed for (magenta in Fig 1, A and B) is fused to the carboxyl terminus of CFP-C (dark blue in Fig. 1, A and B). Competition is quantified as the loss of yellow fluorescence of the CFP-C-β/YFP-N-γ complex upon co-expression of Cer-N-γ subunits (See Fig. 3). Conversely, to compare the abilities of different β subunits to compete for a common γ subunit, one of the β subunits (red in Fig. 1C) is fused to the carboxyl terminus of YFP-N (yellow in Fig. 1C) and each of the β subunits (green in Fig. 1D) is fused to the carboxyl terminus of Cer-N (cyan in Fig. 1D). The γ subunit that is competed for (magenta in Fig. 1, C and D) is fused to the carboxyl terminus of CFP-C (dark blue in Fig. 1, C and D). Competition is quantified as the loss of yellow fluorescence of the CFP-C-γ/YFP-N-β complex upon co-expression of Cer-N-β subunits. Relative effectiveness in competition assays is normalized to the expression levels of the subunits by means of immunoblots using antibodies to GFP that quantify expression of Cer-N-β and Cer-N-γ under the same transfection conditions as the fluorescence measurements.
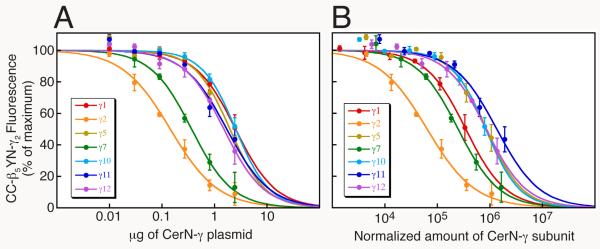
FIG. 3.
β5 interacts preferentially with γ2 rather than γ1, γ5, γ7, γ10, γ11, or γ12. (A) Competition between Cer-N-γ subunits and YFP-N-γ2 for limiting amounts of CFP-C-β5. The intensity of CFP-C-β5YFP-N-γ2 was measured in the presence of each Cer-N-γ subunit or empty vector. HEK-293 cells were transfected with 0.6 μg each of plasmids expressing CFP-C-β5 and YFP-N-γ2, and the indicated μg of each Cer-N-γ plasmid. The total amount of plasmid in each transfection was maintained at 3.63 μg using pcDNAI/Amp. Values represent the means ± S.E. from 3 experiments performed in duplicate. (B) CFP-C-β5YFP-N-γ2 intensity is expressed as a function of the relative amounts of co-expressed Cer-N-γ. Expression levels were determined in HEK-293 cells transfected with 0.6 μg each of plasmids expressing CFP-C-β5 and pcDNAI/Amp, and 0.03, 0.09, 0.27, or 2.43 μg of each Cer-N-γ plasmid. The total amount of plasmid in each transfection was maintained at 3.63 μg using pcDNAI/Amp. The expression levels of the Cer-N-γ subunits varied linearly and the data were fit by linear regressions. The plasmid amounts used in (A) were multiplied by Cer-N-γ expression/μg plasmid to yield the normalized amount of each Cer-N-γ subunit. CC indicates CFP-C and YN indicates YFP-N. (Reprinted from (14) with permission from the American Society for Pharmacology and Experimental Therapeutics and from (18) with permission from Elsevier.)
The interaction preferences of β and γ subunits identified using BiFC most likely indicate association preferences, because BiFC appears to be irreversible (8, 9). As β and γ generally associate irreversibly, this is not a concern. The only reported potential exceptions are β5γ2 (10) and β4γ11 (11), which are unstable in vitro.
Multicolor BiFC can also be applied to visualizing dynamic events involving pairs of βγ complexes using fluorescence microscopy. For example, differences in the kinetics and localization patterns of GPCR-stimulated βγ internalization responses can be visualized and quantified. Such differences may have functional importance in that variability in the rates of agonist-stimulated βγ internalization may cause differences in the deactivation kinetics of plasma membrane-associated effectors. Alternatively, different rates of βγ internalization may lead to different activation rates of effectors located in intracellular compartments.
2. Materials
2.1. Producing fusions of β and γ subunits to fluorescent protein fragments
BiFC vectors: YFP(1-158)/pcDNAI/Amp, Cerulean(1-158)/pcDNAI/Amp, and CFP(159-238)/pcDNAI/Amp (12). These plasmids encode resistance to ampicillin and may be obtained from our laboratory (see Note 1).
cDNAs of β and γ subunits for which BiFC constructs have not been made (see Note 1).
TaqPlus Precision PCR System (Stratagene, La Jolla, CA).
Qiaquick PCR purification and Qiagen MinElute Gel Extraction kits (Qiagen, Valencia, CA).
PCR machine.
2.2. Transient transfections to compare association preferences of β and γ subunits
HEK-293 cells (ATCC, CRL-1573).
Minimal Essential Medium with Earle’s salts with L-glutamine (MEM) (Invitrogen/Life Technologies, #11095-080).
Fetal Bovine Serum (Hyclone, #SH30071.03).
Trypsin-EDTA solution (0.05% trypsin, 0.53 mM EDTA, Invitrogen/Life Technologies, #25300-054).
Lipofectamine 2000 Reagent (Invitrogen/Life Technologies, #11668).
Opti-MEM I Reduced Serum Medium (Invitrogen/Life Technologies, #31985)
60-mm tissue culture dishes
2.3. Measurement of BiFC βγ fluorescence using a spectrofluorometer
PC1 photon-counting spectrofluorometer (ISS, Champaign, IL) or equivalent instrument. The spectrofluorometer is configured with motorized filter wheels on both the excitation path between the excitation monochrometer and the sample, and on the emission path between the sample and the emission monochrometer. The slits on the excitation and emission monochrometers are set to a 16 nm band-pass.
430/25 and 492/18 band-pass filters, 1.3 OD neutral density filter, and 455, 515, and 590 long-pass filters (Chroma, Rockingham, VT).
Glass fluorometer cuvettes with Teflon Covers (Cole-Palmer #H-83200-10).
HBSS+CaCl2 media (20 mM Hepes, pH 7.2, 118 mM NaCl, 4.6 mM KCl, 10 mM D-glucose, 1 mM CaCl2).
HBSS+EDTA media (20 mM Hepes, pH 7.2, 118 mM NaCl, 4.6 mM KCl, 10 mM D-glucose, 0.5 mM EDTA).
2.4. Correcting for the expression levels of Cer-N-β and Cer-N-γ subunits
Dulbecco’s Phosphate-Buffered Saline (D-PBS) with calcium and magnesium (Invitrogen/Life Technologies # 14040).
Dulbecco’s Phosphate-Buffered Saline (D-PBS) without calcium and magnesium (Invitrogen/Life Technologies # 14190).
Coomassie Plus Protein Assay Reagent Kit (Pierce Biotechnology, Rockford, IL).
2 mg/ml Bovine Serum Albumin Standard Ampules (Pierce Biotechnology, Rockford, IL)
SDS-PAGE standards, low range (Bio-Rad Laboratories #161-0304).
XCell SureLock Mini-Cell and XCell II Blot Module Kit CE Mark (Invitrogen/Life Technologies).
Nu-PAGE Bis-Tris gels, NuPAGE MES SDS Running Buffer, Nu-PAGE Antioxidant, NuPAGE LDS Sample Buffer, NuPAGE Transfer Buffer (Invitrogen/Life Technologies).
Dithiothreitol (DTT) (Sigma-Aldrich).
Nitrocellulose, 0.45 μm pore size or Invitrolon PVDF (Invitrogen/Life Technologies).
SuperBlock T20 TBS Blocking Buffer (Pierce Biotechnology, Inc, Rockford, IL).
Rabbit polyclonal antibody to residues 3 to 17 of GFP (Anti-GFP, N-terminal #G1544; Sigma-Aldrich, St. Louis, MO) (used for Cer-N-γ subunits) and goat polyclonal antibody to full-length GFP (#600-101-215, Rockland Immunochemicals, Gilbertsville, PA) (used for Cer-N-β or Cer-N-γ subunits).
Goat anti-rabbit IgG-peroxidase (Sigma-Aldrich #A6154) to use with Anti-GFP, N-terminal antibody, and rabbit anti-goat IgG peroxidase (Sigma-Aldrich #A5420) to use with full-length GFP antibody.
TBS-Tween (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20).
SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology, Rockford, IL).
FluorChem SP Imaging System (Alpha Innotech, San Leandro, CA) or equivalent instrument.
2.5. Imaging dynamic events involving pairs of βγ complexes in living cells
Lab-Tek II, 4 well chambered coverslips (Fisher Scientific).
A white light spinning disc confocal microscope comprised of an Olympus IX81 inverted microscope, UIS2 60x 1.42 N.A. objective, IX2-DSU spinning disc system, 100 watt mercury arc lamp, Hamamatsu C9100-02 electron multiplier camera, Ludl filter wheels, shutters, and xy stage, under the control of IPLab software (BD Biosciences, San Jose, CA), or equivalent fluorescence microscope that can image live cells labeled with CFP, YFP, and mCherry (13).
Excitation and emission filters for CFP (438/24, 483/32), YFP (504/12, 542/27), Red (589/15, 632/22), and a triple dichroic (FF444/521/608) (Semrock, Rochester, NY).
CSMI stage incubator (Harvard Apparatus, Holliston, MA) for imaging at 37°C.
Minimal Essential Medium (MEM) powder with Earle’s salts, with L-glutamine, without sodium bicarbonate (Invitrogen/Life Technologies, #61100-061). To prepare HEPES-buffered MEM, add HEPES to 20 mM and pH to 7.4, then sterilize by filtration.
mCherry-Mem, a membrane marker used for quantifying plasma membrane association of G protein subunits (14) that can be obtained from our lab.
Cintiq pen-based display screen (Wacom, Vancouver, WA).
3. Methods
3.1. Producing fusions of β and γ subunits to fluorescent protein fragments
Using PCR, add a linker sequence encoding Arg-Ser-ILe-Ala-Thr and a BamHI site to the 5′ end of the β or γ subunit cDNA and a Bgl II site to the 3′ end. See Fig. 2 for an example of the coding and noncoding primers used for β1 BiFC constructs (see Note 2).
Digest the PCR product with Bam HI and Bgl II and subclone it into the Bgl II site of one of these BiFC vectors: YFP(1-158)/pcDNAI/Amp, Cerulean(1-158)/pcDNAI/Amp, or CFP(159-238)/pcDNAI/Amp (see Note 3).
FIG. 2.
PCR primers used to produce β1 cDNA for subcloning into BiFC vectors. The human β1 sequence (obtained from Janet Robishaw, Weis Center for Research) was used.
3.2. Transient transfections to compare association preferences of β and γ subunits
For each transfection, plate 1.6 × 106 HEK-293 cells per 60-mm dish in 4 mL of MEM containing 10% FBS. Incubate the cells at 37°C, 5% CO2. Transfections are performed in duplicate and each experiment is repeated at least 3 times.
24 hours later, transfect the cells with BiFC β and γ plasmids. Transfect with a range of plasmid amounts (see Note 4). For each transfection, dispense plasmid into a sterile 1.5 mL microcentrifuge tube. In a sterile hood, add 400 μL of Opti-MEM I to each tube.
In a separate microcentrifuge tube, add 6 μL of Lipofectamine 2000 Reagent to 400 μL of Opti-MEM I. Mix well by inverting the tube several times.
After 5 min, add the Lipofectamine 2000 mixture to the plasmid mixture.
After 20 min, add the 800 μL plasmid-Lipofectamine 2000 mixture to the cells by dripping gently all over the plate. Incubate the cells at 37°C, 5% CO2.
3.3. Measurement of BiFC βγ fluorescence using a spectrofluorometer
Two days after the transient transfections, calibrate the spectrofluorometer as described in the instrument manual.
Make measurements of CFP and YFP fluorescence and of light scattering for each sample. For CFP measurements, the excitation monochrometer is set to 430 nm with a 430/25 band-pass filter, and the emission monochrometer is set to 480 nm with a 455 long-pass filter. For YFP measurements, the excitation is set to 492 nm with a 492/18 band-pass filter, and emission is set to 530 nm with a 515 long-pass filter. The cell density of each sample is determined from a light scattering measurement at 650 nm. Excitation and emission monochrometers are set to 650 nm, and a 1.3 OD neutral density filter in combination with a 590 long-pass filter is used in the excitation filterwheel (see Note 5). Control of the monochrometers, motorized filterwheels, and data acquisition is done using the Vinci software program (ISS).
Run a buffer control using HBSS+EDTA media. Values from this control will be subtracted from all measurements of the cells.
To prepare cell suspensions, add 4 mL of HBSS+CaCl2 media to the dishes, swirl slightly (to get rid of the phenol red in the media), aspirate, and then add 2 mL of HBSS+EDTA media. Scrape the cells off with a rubber policeman, pass through a pipet several times to break up clumps, and suspend in a 1 cm square glass cuvette with a magnetic stir bar. Lightly flick the bottom of the cuvette to get bubbles out of the stir bar area.
Make dilutions of cells transfected with vector alone to produce an autofluorescence versus light scattering standard curve. Make three serial 1:2 dilutions of the cells in HBSS+EDTA media by adding 2 mL of cells to 2 mL of HBSS+EDTA. Measure YFP, CFP, and light scattering for the undiluted, 1:2, 1:4, and 1:8 dilutions. Fit a line to the data.
Measure the YFP, CFP, and light scattering signals of the undiluted experimental samples. Subtract autofluorescence from the YFP and CFP signals of these samples using their light scattering values and the autofluorescence standard curve.
Express the relative preferences of the limiting subunit for the competing subunits as the IC50 for inhibition by the Cer-N-subunits of the yellow fluorescence produced by the CFP-C-subunit/YFP-N-subunit complex. For example, for inhibition of association of YFP-N-γ2 with CFP-C-β5 by Cer-N-γ subunits, the IC50 is defined as μg of Cer-N-γ subunit plasmid that produces a 50% decrease in the intensity of CFP-C-β5YFP-N-γ2. To determine IC50 values, the data are fit to: Y = (100)/(1 + (X/a)b), where X is μg of transfected Cer-N-γ plasmid, Y is the % of maximal fluorescence produced by CFP-C-β5YFP-N-γ2, a is the half-maximal inhibitory concentration (IC50) of the Cer-N-γ subunit, and b is the slope factor. Fig. 3A shows the results of competition between a set of Cer-N-γ subunits with YFP-N-γ2 for association with CFP-C-β5.
3.4. Correcting for the expression levels of Cer-N-β and Cer-N-γ subunits (see Note 6)
Transfect HEK-293 cells with the same amounts of Cer-N-subunits and CFP-C-subunits as in section 3.2, and substitute an equal amount of empty vector for the amount of transfected YFP-N-subunit. Keep the total amount of transfected plasmids constant using empty vector.
2 days later, aspirate media (4 mL) from the 60-mm dishes.
Gently add and remove 4 mL of ice-cold D-PBS with calcium and magnesium.
Add 4 mL of ice-cold D-PBS without calcium or magnesium and dislodge the cells from the dish using a rubber policeman.
Determine protein concentration in 50 μl of cells using Coomassie Plus Protein Assay (Micro Test Tube Protocol) with a standard curve of 0, 2, 5, 10, and 20 μg of Bovine Serum Albumin.
Spin down 15 μg aliquots of cells in refrigerated microcentrifuge and resuspend in 7 μl of D-PBS without calcium or magnesium. Then add 3 μl of NuPAGE sample buffer containing DTT (2.5 μl of NuPAGE sample buffer plus 0.5 μl of 1M DTT). Boil 5 min and run on Nu-PAGE Bis-Tris gel with 0.5 μl of SDS-PAGE standards.
Transfer proteins from gels to Nitrocellulose or Invitrolon PVDF using XCell II Blot Module.
Incubate Nitrocellulose or Invitrolon PVDF with 0.2 % Ponceau S in 3% trichloroacetic acid on shaker for a few minutes, rinse with H2O, mark locations of molecular weight markers with a permanent marker, produce an image of the blot to have a record of the sample protein loadings, and then incubate the blot in SuperBlock T20 TBS Blocking Buffer for 30 minutes with shaking. Replace the Blocking Buffer and shake for another 30 minutes.
For Cer-N-γ subunits, incubate either with anti-GFP, N-terminal antibody at a dilution of 1:2,500, or with full-length GFP antibody at a dilution of 1:400 in TBS-Tween overnight on a shaker at 4°C. For Cer-N-β subunits, best results are obtained with the full-length GFP antibody at a dilution of 1:400.
For blots incubated in full-length GFP antibody, incubate for 1 hour at room temperature in anti-goat IgG peroxidase at a dilution of 1:40,000 in TBS-Tween. For blots incubated in anti-GFP, N-terminal antibody, incubate for 1 hour at room temperature in anti-rabbit IgG-peroxidase at a dilution of 1:2,000.
Detect antigen-antibody complexes using SuperSignal West Pico Chemiluminescent Substrate and a FluorChem SP Imaging System or equivalent instrument.
Quantify expression levels using of IPLab software (BD Biosciences, San Jose, CA) or equivalent imaging program.
Normalize the IC50 values calculated in section 3.3.7 by multiplying these values (in μg of Cer-N-subunit plasmid) by Cer-N-subunit expression/μg plasmid to yield the normalized amount of each Cer-N-subunit (see Fig. 3b).
3.5. Imaging dynamic events involving pairs of βγ complexes in living cells
Plate HEK-293 cells at a density of 105 cells per well in 500 μL of MEM on Lab-Tek II, 4 well chambered coverslips.
24 hours later, transiently transfect the cells with plasmids encoding the CFP-C-subunit, YFP-N-subunit, and Cer-N-subunit of interest, along with mCherry-Mem, using 0.25 μL of Lipofectamine 2000 Reagent. Co-expressing plasmids encoding a GPCR and an associated α subunit enables investigations of agonist-stimulated internalization of βγ complexes. The transfection procedure is the same as in section 3.2 except that the plasmids and the Lipofectamine 2000 reagent are each suspended in 50 μL of Opti-MEM I (see Note 7).
2 days after the transfection, image the cells at 60x using an Olympus white light spinning-disc confocal microscope or equivalent instrument. At least one hour before imaging, replace the bicarbonate-buffered medium with HEPES-buffered MEM (see Note 8).
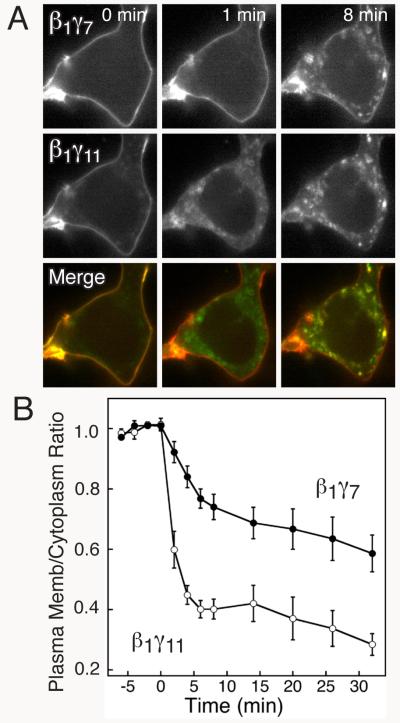
Collect images of cells in the CFP, YFP, Red, and DIC channels at timed intervals before and after stimulation of a GPCR with an agonist. A motorized x-y-z stage makes it possible to collect images of cells located at 5-6 independent positions in the well during a single experiment. Cells selected for imaging should express all of the labeled proteins and have a clearly delineated plasma membrane. Individual exposure times should be optimized for each cell and color channel. Fig. 4A shows images of β1γ7 and β1γ11 internalizing from the plasma membrane in response to stimulation of the β2-adrenergic receptor.
Determine the plasma membrane to cytoplasm intensity ratios for the βγ complexes at each time point. Make a “high resolution” version of the mCherry-Mem image that has a flat background and peaks corresponding to in-focus features. First, blur the image with a 15×15 pixel filter to produce a “low resolution” image. Then, subtract the low resolution image from the original image, followed by a 5×5 pixel filter to smooth noise. Threshold the high resolution image with an intensity cut-off value that selects pixels that match the features seen in the unprocessed image. Use the same threshold value for all time points. Draw a border around the edge of the cell centered on the plasma membrane, 6-10 pixels wide (0.6 – 1.0 μm) using a Cintiq pen-based display screen. Pixels within this border that are above the threshold are counted as plasma membrane pixels. Also, circle the nucleus of each cell in the DIC image. The cytoplasm pixels are inside the border centered on the plasma membrane and outside of the nucleus. Determine the average intensities of the plasma membrane and cytoplasm pixels for the CFP-C-subunit/YFP-N-subunit complex, the CFP-C-subunit/Cer-N-subunit complex, and mCherry-Mem in the original YFP, CFP, and red images, respectively. Divide the plasma membrane to cytoplasm intensity ratios of the βγ complexes by those of mCherry-Mem (normalized to a value of 1 for the first time point) (see Note 9). Fig. 4B shows quantification of the internalization responses of β1γ7 and β1γ11 upon stimulation of the β2-adrenergic receptor.
FIG. 4.
The stimulus-induced localization pattern of β1γ11 differs from that of β1γ7. (A) YFP (top), CFP (middle), and merge (bottom) images from the same cell expressing CFP-C-β1YFP-N-γ7 and CFP-C-β1Cer-N-γ11 acquired at the indicated times before and after stimulation with 10 μM isoproterenol. In the merge image, co-localization of CFP-C-β1YFP-N-γ7 (red) and CFP-C-β1Cer-N-γ11 (green) is indicated in yellow. (Reprinted from (18) with permission from Elsevier.) (B) Quantification of isoproterenol-stimulated decreases in plasma membrane/cytoplasm ratios of CFP-C-β1YFP-N-γ11 and CFP-C-β1Cer-N-γ7 in HEK-293 cells. The cells were stimulated with 10 μM isoproterenol immediately after time = 0. Values represent the mean ± S.E of measurements in 7 cells.
Footnotes
Our laboratory has produced a wide assortment of BiFC β and γ subunit constructs (12, 14, 15). These constructs and BiFC vectors can be obtained by sending a request by E-mail to Catherine Berlot (chberlot@geisinger.edu).
The BiFC vectors, YFP(1-158)/pcDNAI/Amp, Cerulean(1-158)/pcDNAI/Amp, and CFP(159-238)/pcDNAI/Amp fuse the fluorescent protein fragment to the amino terminus of the β or γ subunit.
This strategy requires that the β or γ subunit cDNA does not have internal Bam HI or Bgl II sites. If these sites are present, they will need to be removed using silent mutations that do not change the amino acid sequence (see Fig. 2). cDNAs digested with Bam HI and Bgl II have compatible sticky ends that when ligated do not regenerate either site (see Fig. 2). With this strategy, the fusion protein cDNAs can be moved to different vectors as Bam HI/ Bgl II cassettes.
In competition experiments, the subunit that is competed for is expressed in a limiting amount compared to the competing subunits. Loss of YFP fluorescence from CFP-C-β/YFP-N-γ or YFP-N-β/CFP-C-γ complexes is measured in the presence of a range of amounts of Cer-N-γ or Cer-N-β subunits, respectively. The optimum amounts of plasmids to transfect need to be determined empirically by measuring competition between fusions of Cer-N and YFP-N to the same subunit. The initial fluorescence of the CFP-C-β/YFP-N-γ or YFP-N-β/CFP-C-γ complex needs to be high enough to provide a workable dynamic range of fluorescence intensities in the presence of the competing Cer-N-γ or Cer-N-β subunits. The β subunit fusions generally express at lower levels than do the γ subunit fusions, so optimal conditions for measuring competition of γ subunits for β subunits may differ from those for competition of β subunits for γ subunits. For example, to compare γ subunits competing for β1 or β5, HEK-293 cells were transfected with 0.6 μg each of plasmids expressing CFP-C-β1 or CFP-C-β5 and YFP-N-γ2 and 0, 0.01, 0.03, 0.09, 0.27, 0.81, or 2.43 μg of each Cer-N-γ subunit (14, 15) (Fig. 3). The total amount of plasmid was maintained at 3.63 μg by making up the difference with empty vector (pcDNAI/Amp). The total amount of transfected plasmid needs to be constant, because promoter competition decreases fluorescence. In contrast, to compare β1 and β5 competing for γ2, cells were transfected with 0.3 μg of CFP-C-γ2 plasmid, 0.6 μg of YFP-N-β1 plasmid, and 0.033, 0.1, 0.3, 0.9, 2.7, or 8.1 μg of plasmids encoding either Cer-N-β1 or Cer-N-β5. The total amount of plasmid was maintained at 9 μg using pcDNAI/Amp (14).
Two significant sources of background signal must be eliminated in order to measure fluorescent proteins in a suspension of cells accurately. One source of background signal is the strong light scattering property of cells. The monochrometers found in most spectrofluorometers transmit a small amount of stray light outside the selected wavelength band. The band-pass filters commonly used for fluorescence microscopy block significantly more stray light. A band-pass filter between the excitation monochrometer and the sample will block any stray light in the emission wavelength range that would be scattered and detected. Additionally, a band-pass or long-pass filter between the sample and the emission monochrometer will prevent scattered excitation light from reaching the emission monochrometer. With filters in place, the signal from a nonfluorescent scattering sample, such as a dilute solution of glass beads (glass-milk) should be the same as that of a buffer control. A second background signal is autofluorescence from cellular proteins. Autofluorescence is proportional to cell density, which is determined with the described light scattering measurement.
In addition to determining the expression levels of BiFC β and γ constructs, it is important to assess the functionality of BiFC βγ complexes. We have demonstrated that YFP-N-β1YFP-C-γ complexes potentiate αs-mediated activation of adenylyl cyclase in COS-7 cells (12) and that CFP-C-β5Cer-N-γ complexes activate phospholipase C-β2 expressed in HEK-293 cells (14).
The optimal amounts of transfected plasmids need to be determined empirically. For instance, when dually imaging CFP-C-β1YFP-N-γ7 and CFP-C-β1Cer-N-γ11 for Fig. 4, more YFP-N-γ7 than Cer-N-γ11 plasmid was used to normalize the fluorescence intensities of the two βγ complexes. For these images, HEK-293 cells were transfected with the following amounts (in μg) of plasmids: αs, 0.3; CFP-C-β1, 0.3; YFP-N-γ7, 0.1125; Cer-N-γ11, 0.0375; β2AR, 0.1; and mCherry-Mem, 0.0025. Because CFP-C-subunit/Cer-N-subunit complexes are brighter than CFP-C-subunit/YFP-N-subunit complexes, it may be helpful to use a Cer-N fusion to the subunit with the lower expression level.
It is important to replace bicarbonate-buffered medium with HEPES-buffered medium to keep the pH constant when viewing cells in the room environment, because changes in pH can alter localization patterns. Alternatively, the dish can be kept in a 5% CO2 atmosphere while imaging.
Changes in cell shape during time courses will alter the membrane to cytoplasm ratio. This is corrected for with the mCherry-Mem measurements, because the distribution of the membrane marker does not change in response to agonist stimulation. A bleach correction is not necessary because ratios of fluorescence are calculated.
5. References
- 1.Robishaw JD, Berlot CH. Translating G protein subunit diversity into functional specificity. Curr Opin Cell Biol. 2004;16:206–209. doi: 10.1016/j.ceb.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Mullah B, Hansen C, Asundi J, Robishaw JD. Ribozyme-mediated suppression of the G protein gamma7 subunit suggests a role in hormone regulation of adenylylcyclase activity. J Biol Chem. 1997;272:26040–8. doi: 10.1074/jbc.272.41.26040. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Mullah BK, Robishaw JD. Ribozyme approach identifies a functional association between the G protein beta1gamma7 subunits in the beta-adrenergic receptor signaling pathway. J Biol Chem. 1999;274:17365–71. doi: 10.1074/jbc.274.24.17365. [DOI] [PubMed] [Google Scholar]
- 4.Schwindinger WF, Betz KS, Giger KE, Sabol A, Bronson SK, Robishaw JD. Loss of G protein gamma 7 alters behavior and reduces striatal alpha(olf) level and cAMP production. J Biol Chem. 2003;278:6575–9. doi: 10.1074/jbc.M211132200. [DOI] [PubMed] [Google Scholar]
- 5.Schwindinger WF, Giger KE, Betz KS, Stauffer AM, Sunderlin EM, Sim-Selley LJ, Selley DE, Bronson SK, Robishaw JD. Mice with deficiency of G protein gamma3 are lean and have seizures. Mol Cell Biol. 2004;24:7758–68. doi: 10.1128/MCB.24.17.7758-7768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–45. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–9. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 8.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7:449–56. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005;127:146–57. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 10.Jones MB, Garrison JC. Instability of the G-protein beta5 subunit in detergent. Anal Biochem. 1999;268:126–33. doi: 10.1006/abio.1998.3064. [DOI] [PubMed] [Google Scholar]
- 11.McIntire WE, MacCleery G, Murphree LJ, Kerchner KR, Linden J, Garrison JC. Influence of differential stability of G protein betagamma dimers containing the gamma11 subunit on functional activity at the M1 muscarinic receptor, A1 adenosine receptor, and phospholipase C-beta. Biochemistry. 2006;45:11616–31. doi: 10.1021/bi0604882. [DOI] [PubMed] [Google Scholar]
- 12.Hynes TR, Tang L, Mervine SM, Sabo JL, Yost EA, Devreotes PN, Berlot CH. Visualization of G protein betagamma dimers using bimolecular fluorescence complementation demonstrates roles for both beta and gamma in subcellular targeting. J Biol Chem. 2004;279:30279–86. doi: 10.1074/jbc.M401432200. [DOI] [PubMed] [Google Scholar]
- 13.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 14.Yost EA, Mervine SM, Sabo JL, Hynes TR, Berlot CH. Live Cell Analysis of G Protein beta5 Complex Formation, Function, and Targeting. Mol Pharmacol. 2007;72:812–25. doi: 10.1124/mol.107.038075. [DOI] [PubMed] [Google Scholar]
- 15.Mervine SM, Yost EA, Sabo JL, Hynes TR, Berlot CH. Analysis of G Protein beta{gamma} Dimer Formation in Live Cells Using Multicolor Bimolecular Fluorescence Complementation Demonstrates Preferences of beta1 for Particular {gamma} Subunits. Mol Pharmacol. 2006;70:194–205. doi: 10.1124/mol.106.022616. [DOI] [PubMed] [Google Scholar]
- 16.Ormo M, Cubbitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequoria victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 17.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–9. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 18.Hynes TR, Yost E, Mervine S, Berlot CH. Multicolor BiFC analysis of competition among G protein beta and gamma subunit interactions. Methods. 2008;45:207–13. doi: 10.1016/j.ymeth.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]