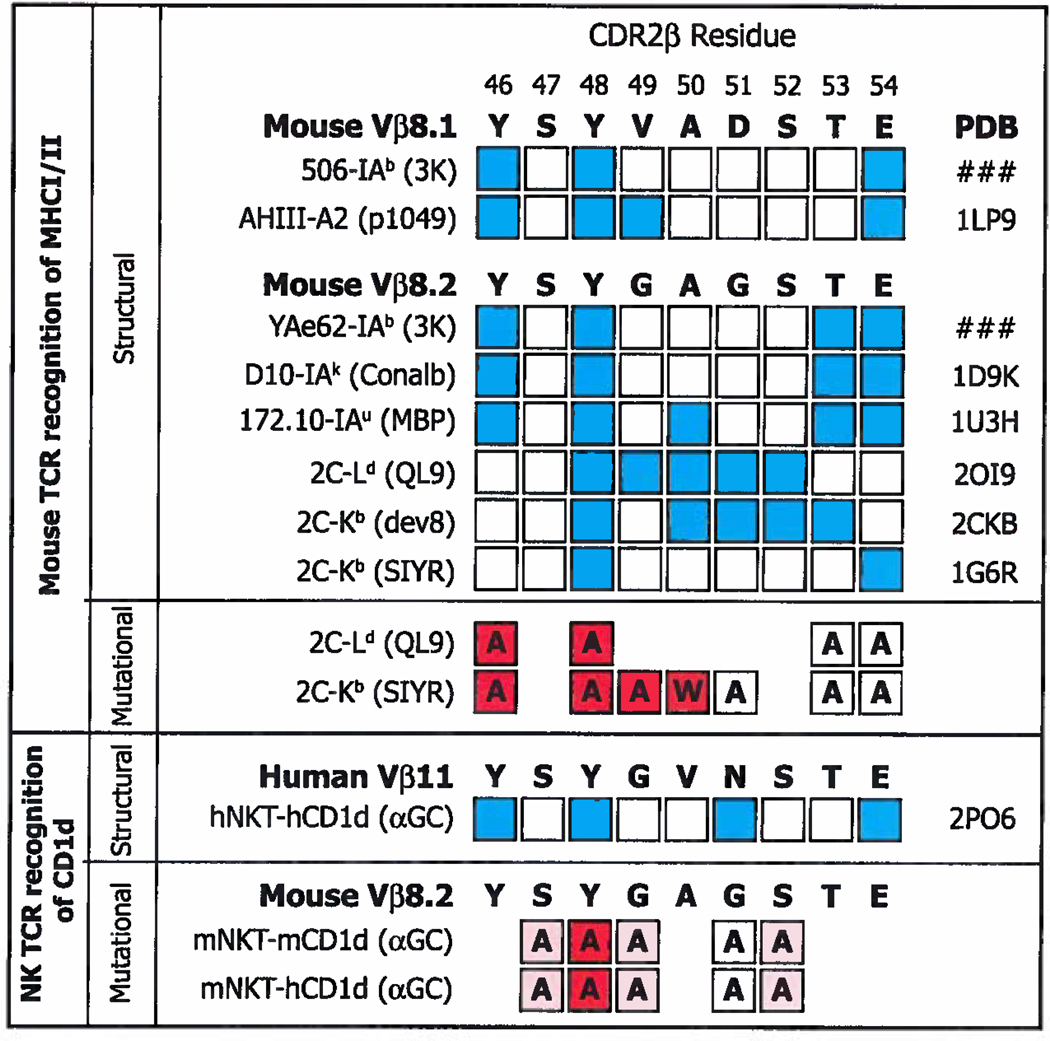
Figure 10. Some of the CDR2β amino acids used to contact classical MHCI/MHCII and CD1d are identical.
Above. The amino acid sequences of mVβ8.1 and mVβ8.2 in bold. Amino acids that have been shown in sturtures to bind MHC are indicated in blue, together with the names of the TCRs and pdb files used. Amino acids in white squares do not contact MHC in the indicated structure. Below this are shown the results of mutational analyses, in which the amino acid in question was mutated as shown in each square. The influence of each mutation is indicated by particular shades of red, red filled squares indicate >1 log change in reactivity, while white indicates a <0.5 change in log reactivity (122, 123). Below Contact points identified by structural analysis (68) between a hVα24i TCR and hCD1d/αGC and identified by mutational analyses, using staining with CD1d/αGC tetramers, between a mVα14i TCR and mCD1d/αGC or hCD1d/αGC (Scott-Browns et al. submitted for publication). Red filled squares indicate >50% loss in tetramer mean fluorescent intensity (MFI), pink indicates a change between 10% and 50% change in tetramer MFI, while white indicates a <10% change in tetramer MFI.