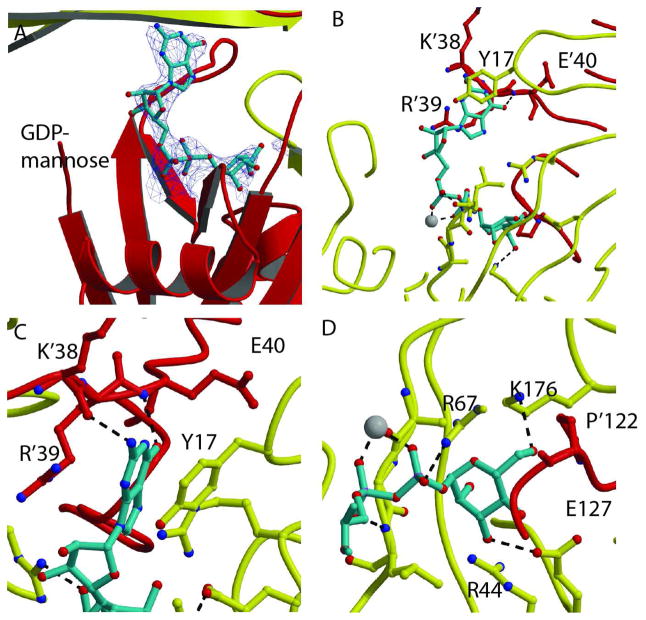
Fig. 3. Recognition of GDP-mannose.
A. Electron density omit σA map of the GDP-mannose in the active site of the GDPMK E100A mutant (contour level =1σ). Red and yellow ribbons show the two monomers. The magnesium ion is shown as a white sphere. B. GDP-mannose is shown as turquoise sticks. The residues of both chains that form the binding pocket are shown as sticks. Hydrogen bonds to both the guanosine base and the mannose are shown as black dashes. C. Guanine bound to GDPMK by π- π interactions to R’39 and Y’17 of the opposite monomer. The guanine base is further stabilized by hydrogen bonds to K’38 and E’40. D. GDPMK recognition of mannose mediated by hydrogen bonds to conserved E127 and to K176, a residue that is not found in the ADP-ribose pyrophosphatase family.