Figure 2.
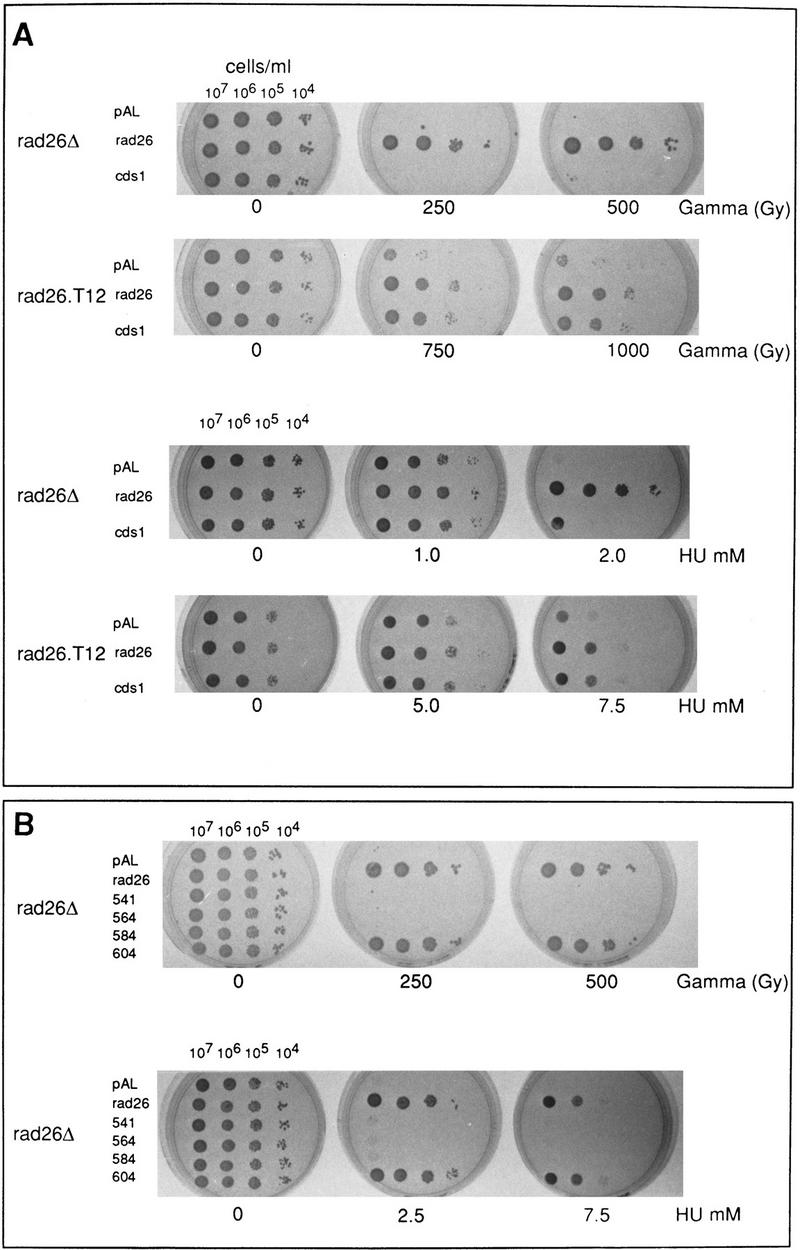
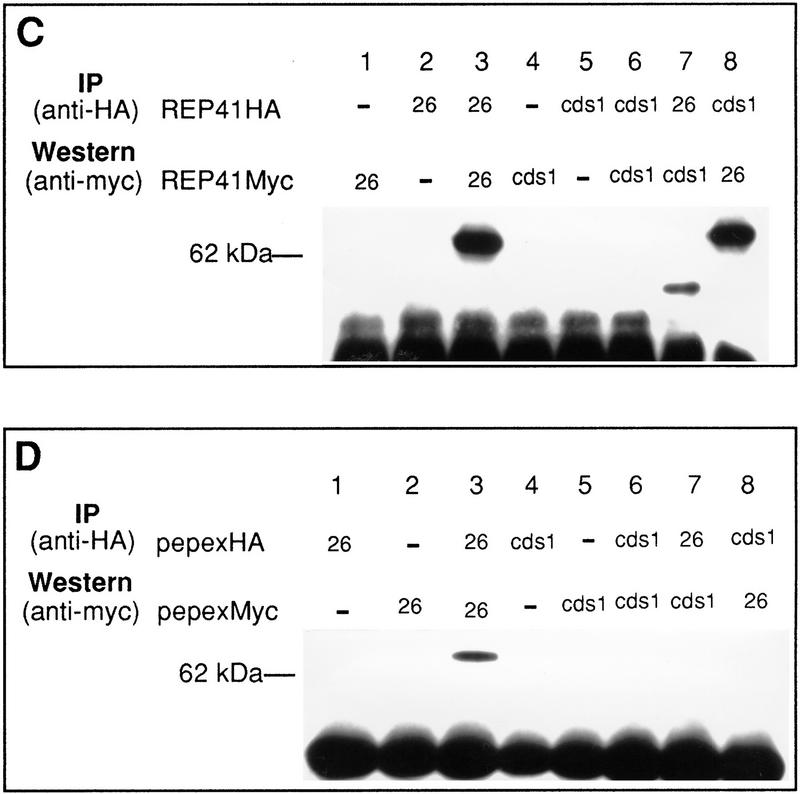
cds1 is a multicopy suppresser of rad26.T12 mutant cells. (A) rad26 null and rad26.T12 mutant cells were transformed with rad26 genomic DNA, cds1 genomic DNA, or an empty vector control (pAL). Cells were serially diluted (from 107 cells/ml to 104 cells/ml), spotted onto plates and irradiated at the doses indicated. Cells were also spotted onto plates containing a range of HU concentrations. (B) rad26 null cells were transformed with the rad26 genomic clone, the empty vector (pAL) or rad26 constructs containing carboxy-terminal deletions. Numbers correspond to the length of the constructs (amino acids). Full-length Rad26 is 614 amino acids. Cells were diluted to the concentrations shown and either spotted onto plates and irradiated at the doses indicated or spotted onto plates containing different concentrations of HU. (C) Cds1 immunoprecipitates with Rad26 in S. pombe extracts. Wild-type S. pombe was transformed with REP41 (leucine selection) and REP42 (uracil selection) plasmids containing cds1 and rad26 cDNA tagged with either HA or Myc epitopes. Immunoprecipitation was carried out on extracts from logarithmically growing cells, after 18 hr growth without thiamine. Immunocomplexes were processed for Western blotting with anti-Myc antibody. Rad26 associates with itself (lane 3) and with Cds1 (lanes 7,8). The empty vector (lanes 1,2,4,5) was used as control. We estimate overexpression at ∼50× of wild-type protein levels. Comparing the amount of Cds1 precipitated with Rad26 to total protein, we estimate ∼3%–5% recovery. (D) Immunoprecipitations were repeated by use of proteins expressed in rabbit reticulocyte lysate. In this case, Rad26 was seen to associate with itself, but not with Cds1, suggesting bridging proteins or posttranslational modification are required for the Rad26–Cds1 interaction. This would be consistent with levels of recovery.