Figure 1. In vitro selection for ZFN-mediated cleavage.
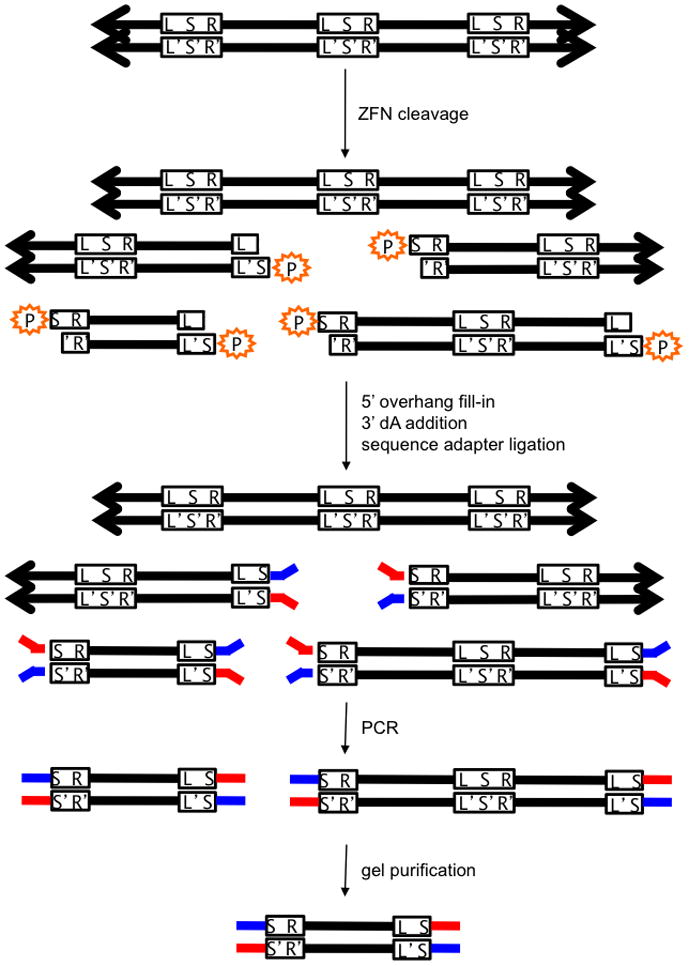
Pre-selection library members are concatemers (represented by arrows) of identical ZFN target sites lacking 5′ phosphates (orange). L = left half-site; R = right half-site, S = spacer; L′, S′, R′ = complementary sequences to L, S, R. ZFN cleavage reveals a 5′ phosphate, which is required for sequencing adapter (red and blue) ligation. The only sequences that can be amplified by PCR using primers complementary to the red and blue adapters are sequences that have been cleaved twice and have adapters on both ends. DNA cleaved at adjacent sites are purified by gel electrophoresis and sequenced. A computational screening step after sequencing ensures that the filled-in spacer sequences (S and S′) are complementary and therefore from the same molecule.
