Abstract
Formation of isoaspartic acid (isoAsp) is a common modification of aspartic acid (Asp) or asparagine (Asn) residue in proteins. Differentiation of isoAsp and Asp residues is a challenging task owing to their similar properties and identical molecular mass. It was recently shown that they can be differentiated using ion-electron or ion-ion interaction fragmentation methods (ExD), as these methods provide diagnostic fragments c + 57 and z• − 57 specific to the isoAsp residue. To date, however, the presence of such fragments has not been explored on peptides with an N-terminal isoAsp residue. To address this question, several N-terminal isoAsp-containing peptides were analyzed using ExD methods alone or combined with chromatography. A diagnostic fragment [M + 2H − 74]+• was observed for the doubly charged precursor ions with N-terminal isoAsp residues. For some peptides, identification of the N-terminal isoAsp residue was challenging due to the low diagnostic ion peak intensity and the presence of interfering peaks. Supplemental activation was used to improve diagnostic ion detection. Further, N-terminal acetylation was offered as a means to overcome the interference problem by shifting the diagnostic fragment peak to [M + 2H − 116]+•.
Introduction
Isoaspartic acid (isoAsp) is an isomer of aspartic acid (Asp) that can be formed by either isomerization of an Asp or deamidation of an asparagine (Asn) residue. Under physiological conditions, both reactions proceed via formation of a cyclic succinimide intermediate followed by rapid hydrolysis to form a mixture of Asp and isoAsp, typically in a 1:3 ratio in short unstructured peptides.1, 2 In proteins, the ratio could be different from 1:3 and influenced by structure.3, 4 The succinimide formation from Asp is ~ 10 – 40 times slower than from Asn at neutral pH, yet the reaction rate may vary greatly depending on the adjacent residues, protein conformation and the proximity of the Asp/Asn residue to the protein surface.1, 5, 6
Asn deamidation and Asp isomerization are common in vivo, particularly in long-lived proteins, which are often associated with aging,7 eye lens abnormalities,8 and amyloid diseases such as Alzheimer disease.9, 10 These modifications may alter protein conformation, activity and stability, and trigger protein aggregation. For example, the 3-dimensional crystal structure of Ribonuclease U2B revealed that formation of isoAsp32 led to a single turn unfolding of the α-helix to form a U-shape loop structure, affecting the hydrolytic activity of the protein.11 Single Asp isomerization was shown to deactivate the antigen-binding region of the immunoglobulin gamma (IgG)-2 antibody.12 The presence of the isomerized Asp may also affect the proteolytic stability of the protein. For example, Lys-C proteolysis is hindered when the Lys residue is adjacent to an isoAsp in IgG-1 antibody;13 isoAsp residues are also resistant to Asp-N proteolysis.14–16 On the other hand, carboxypeptidase Y cleaves N-terminally to an isoAsp residue recognizing its α-carboxylic acid as if it is a carboxyl-terminal amino acid.17
Although in vivo Asn deamidation is irreversible, the isoAsp accumulation can be minimized by protein L-isoaspartyl O-methyltransferase (PIMT),10 which catalyses isoAsp conversion to Asp. Asn deamidation and Asp isomerization are known to play a role in apoptosis18 and have been proposed to serve as a molecular timer of biological events.6 Furthermore, PIMT-mediated isoAsp-Asp interconversion may regulate synaptic transmission in presynaptic proteins.18 In vitro Asp isomerization and Asn deamidation can also occur during protein production and storage, where the PIMT enzyme is not available and the isoAsp products accumulate with time. IsoAsp buildup could be detrimental to protein structural integrity and stability, which affects the shelf-life and potency of the therapeutic monoclonal antibodies (mAb) in pharmaceutical industry as well as all other protein based new drug entities. To avoid undesirable consequences due to drug degradation, such modifications have to be carefully monitored.
A number of techniques have been developed to detect isoAsp, and they are often used in conjunction. These include the Edman degradation reaction,19 PIMT-utilizing assays,20 affinity enrichment of isoAsp-containing proteins,21 use of isoAsp-specific antibodies,10, 22 endoproteinase Asp-N-based approaches,12, 14, 23 isotopic 18O labeling,4, 24, 25 and various liquid chromatography (LC)-based techniques: hydrophobic interaction chromatography,26 size-exclusion high performance liquid chromatography (HPLC),12 cation exchange HPLC,26 Reversed-Phase (RP) HPLC,27, 28 and Ultra HPLC (UPLC).29 Asp/isoAsp differentiation has been demonstrated by tandem mass spectrometric approaches including collisionally activated dissociation (CAD),30, 31 fast atom bombardment,32 and post-source decay fragmentation.33 Recently, isoAsp identification and quantitation by tandem MS methods employing ion-electron or ion-ion interactions (ExD) have been developed34–39 and successfully applied,23, 40, 41 based on the diagnostic fragment ions c + 57 and z• − 57 that result from the Cα − Cβ bond cleavage unique to isoAsp residues. All the above-mentioned MS methods were accomplished in the positive ion mode; however, in the negative ion mode, the isoAsp-containing peptides were not differentiated.42
In spite of a large number of existing approaches, isoAsp identification remains a challenging task. In HPLC, the elution order of Asp/isoAsp-containing peptides depends on conditions of separation (column type, mobile phase, temperature, etc.), materials, and the instrument used.23, 28 In particular, as was previously shown,29, 43 the elution order can be inverted when isoAsp is located at the N-terminus, leading to erroneous assignment. Thus, unless a standard mixture of the same peptide pair separated at the same condition is available, a further analysis would be required for correct isomer assignment. ExD-based tandem MS seems to be a fast and accurate approach well suited for this task. However, to the best of our knowledge, a detailed ExD study of peptides with N-terminally located isoAsp residues has not been performed and the N-terminal isoAsp-specific fragments have not yet been demonstrated. In this work, the potential of ExD for N-terminal isoAsp residue identification was investigated, either by performing the ExD analysis alone or following RP-HPLC separation. The role of N-terminal acetylation and the effect of ion activation in isomer differentiation were also examined.
Experimental
Peptides and reagents
Angiotensin II (Ang II) peptide variant iDRVYIHPF (hereinafter iD represents isoAsp in peptide sequences), synthetic peptides Ac-DGVGDVGGVH-NH2 and Ac-iDGVGiDVGGVH-NH2, and Amyloid beta 1–10 peptide variant NAEFRHNSGY with Asn residues at position 1 and 7 (hereafter Aβ1–10 (N1N7)) were custom synthesized by Peptide 2.0 (Chantilly, VA, USA). Ang II (DRVYIHPF) and Aβ1-10 (DAEFRHDSGY) were ordered from AnaSpec (San Jose, CA, USA). Formic Acid (FA) was purchased from Thermo Scientific (Rockford, IL, USA). HPLC grade Acetonitrile (ACN) and Methanol (MeOH) were obtained from Fisher Scientific (Fair Lawn, NJ, USA). Deionized water was purified by a Millipore Milli-Q Gradient system (R=18.2 MΩ cm and TOC = 9 – 12 ppb) (Billerica, MA, USA). Ammonium Bicarbonate (ABC) was ordered from Sigma-Aldrich (St. Louis, MO, USA).
Deamidation
The Aβ1–10 peptide variant (NAEFRHNSGY) was incubated at 0.44 mM concentration in 0.1 mM ABC, at pH ~7.7 and 37 °C for 4 days. The resulting peptide mixture was separated on reversed-phase C18 column prior to MS/MS analysis.
Chromatography
RP-HPLC peptide separations were performed on an Agilent 1200 Series system (Agilent Technologies, Wilmington, DE, USA) using a reversed-phase C18 column Vydac 218TP5215 (150 × 2.1 mm, 3 µm particles, 300 Å pore size). 4 nmol of peptide mixture (in 20 µl) was injected directly into the column and eluted using a linear gradient of acetonitrile with 0.1% FA (0 min - 1% B, 20 min - 11% B) at 0.7 ml/min flow rate at 60 °C. The chromatograms were measured using UV detection at 214 nm. Fractions were collected and refrigerated at 4 °C prior to MS/MS analysis.
Mass Spectrometry
ExD analyses were performed on:
1) a solariX FTICR instrument (Bruker Daltonics, Billerica, MA, USA) with a 12 T actively shielded magnet: electron capture dissociation (ECD), hotECD, and electron transfer dissociation (ETD). Peptides were nanosprayed with 0.5 – 2 µM concentration in 50:50 ACN:H2O with 0.1% FA following HPLC separation with off-line fraction collection or in 50:50 MeOH:H2O with 0.1% FA when infused directly. Doubly-charged molecular ions were isolated and irradiated with low- (cathode bias 0.4–0.8 V) or high- (cathode bias 4.5 V (hotECD)) energy electrons for 0.05 – 0.5 ms to produce fragments. Each ExD spectrum was the result of 100 scans.
2) an LTQ-Orbitrap XL with ETD capability (Thermo Scientific, San Jose, CA, USA): ETD with or without supplemental activation (SA). Peptides were nanosprayed in solutions as stated above at ~ 0.5 µM using a robotic Nanomate source (Advion, Ithaca, NY, USA). Activation energy parameter “En” of 5 or 15 was applied when SA was on (typical En values range from 0 – 20).
and 3) an amaZon Ion Trap instrument (Bruker Daltonics, Billerica, MA, USA): ETD with or without smart decomposition (SD). Peptides were electrosprayed (2 µL/min, glass capillary temperature 220 °C) at 1 µM concentration in 50:50 MeOH:H2O with 0.1–1% formic acid using an Apollo II ion source.
Fluoranthene was used as the anion radical reagent in all ETD experiments with 150 – 250 ms reagent accumulation, and 50 – 150 ms reaction time was used to produce fragments. Data acquired on solariX and amaZon were analyzed using Bruker’s ESI Compass DataAnalysis 4.0 software. The Orbitrap data were analyzed using Thermo's Xcalibur 2.0.7 software.
Results and Discussion
Diagnostic fragments for the N-terminal Asp/isoAsp
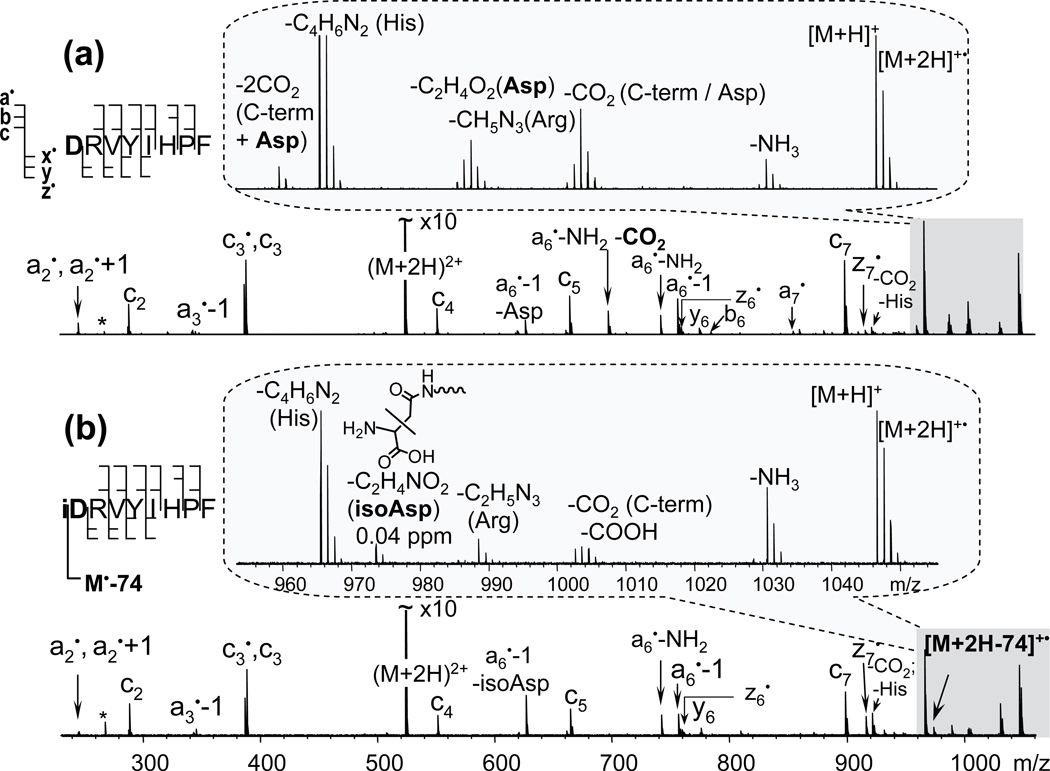
As originally proposed, ECD of peptides with isoAsp located at the nth position may generate a unique complementary ion pair cn-1 + 57 and zm- n + 1 • − 57 as a result of the Cα − Cβ bond cleavage, where m is the total number of amino acid residues and 57, or more accurately, 56.9976 corresponds to the mass of a C2HO2• group. For an isoAsp residue located at the N-terminus, n = 1, and the diagnostic fragments become c0 + 57 and zm• − 57. The c0 fragment is essentially an ammonia molecule which is neither informative nor observed in ECD spectra, and the zm• ion contains no sequence information, and is usually assigned as a loss of ammonia from the charge-reduced species [M + 2H − NH3]+•. Thus, when an N-terminal isoAsp residue is present, the C-terminal diagnostic fragment would be [M + 2H − (NH3 + C2HO2•)]+• or [M + 2H −74.0242]+• (hereinafter indicated as [M + 2H − 74]+•), and the N-terminal diagnostic fragment would be C2H5NO2+• (m/z = 75.032). Although the m/z = 75 peak is usually below the low mass cut-off, it is still possible to look for the corresponding neutral loss [M + 2H − 74]+•.
In order to see if the isoAsp can be differentiated from the Asp when located at the N-terminus, ECD spectra of the two synthetic isomers of Ang II were directly compared (Figure 1). The two spectra were very similar with only a few exceptions. Careful examination revealed the presence of the specific fragment “z8•” − 57 or [M + 2H − 74]+• only in the ECD spectrum of the isoAsp-containing Ang II peptide. In contrast, the loss of 60.0211 (C2H4O2) and a double CO2 loss were observed only for the Asp peptide. Indeed, the loss of 60 is characteristic of the Asp side chain44 and cannot be formed from the isoAsp. The loss of two CO2 molecules can be generated by a combined loss from the C-terminus and from the Asp side chain. Although the CO2 loss can also come from an isoAsp residue, it is normally of a much lower abundance37, 41 or undetectable.38 For example, the a6•− NH2 − CO2 peak was very abundant in the ECD spectrum of the Ang II peptide, but was not detected in that of its isoAsp variant. Thus, in Figure 1a, the CO2 loss from the charge-reduced species was probably generated from the C-terminus.
Figure 1.
ECD spectra of the Angiotensin II (a) and its modified variant with isoAsp at the N-terminus (b). Insets show side-chain losses from the charge-reduced species [M + 2H]+•. IsoAsp specific fragment [M + 2H − 74]+•, produced as a result of the Cα − Cβ bond cleavage within the N-terminal isoAsp residue, is present in the modified variant only, (b) inset.
Analysis of the deamidation products of Aβ1–10 (N1N7) by off-line RP-HPLC followed by ECD
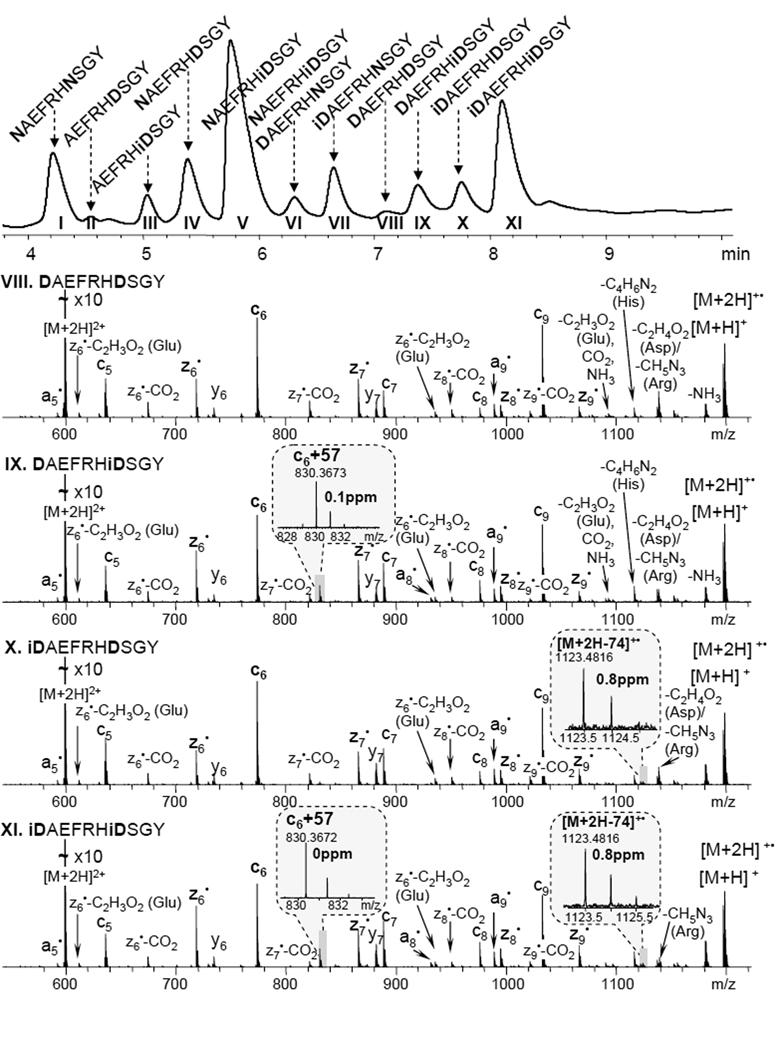
An Aβ1–10 peptide variant with Asn residues at position 1 and 7 was deamidated to artificially introduce isoAsp residues into the sequence. The resulting mixture of peptides was separated on a C18 reversed-phase column. 11 peaks were detected in the chromatogram (figure 2, top panel), collected into fractions, and further analyzed using ECD. It was possible to differentiate and to define the elution order of the isomeric peptides containing isoAsp residues based on the diagnostic fragments observed, i.e. [M + 2H − 74]+• for the isoAsp1 and c6 + 57 for the isoAsp7, as shown in figure 2. Non-deamidated Asn residues were distinguished based on the 0.984 mass difference of the precursor ions and their fragments containing Asn, as shown in the example of z6• fragments (Figure S1. fractions I and V). Fraction VI contained both peptides DAEFRHNSGY and NAEFRHiDSGY, which are isomers and thus indistinguishable at the MS level yet could be easily identified by ECD because some fragment ions exhibited two components, separated by 0.984 Da (Figure S1. fraction VI, inset). The DAEFRHNSGY peptide comprises only a small fraction of fraction VI, based on the relative abundance of the two components within the z6• isotopic cluster. This is consistent with the fact that the DAEFRHNSGY peptide has a lower probability of formation comparing to the NAEFRHiDSGY peptide (from original peptide NAEFRHNSGY) because the Asn in the NA sequence has a slower deamidation rate than Asn in the NS sequence.45, 46 Although the separation of this closely related mixture of peptides was challenging and for some peaks there was no base line separation, all peptides were successfully identified by ECD, highlighting the utility of ECD in isoAspartomic research.
Figure 2.
Chromatogram of the peptide mixture, generated by partial deamidation of the Aβ1–10 (N1N7) peptide variant (top panel). Each chromatographic peak was labeled based on ECD results. The 4 ECD spectra shown correspond to the VIII–XI fractions with the isoAsp specific fragments indicated in the insets. Additional spectra for fractions I–VII are available in supplemental material, figure S1.
Peak interference
Caution should be taken if the peptide of interest contains methionine (Met) or isoleucine (Ile) residues because they may produce side-chain loss fragment ions with m/z close to that of the diagnostic fragment ion: the Met side-chain loss (C3H6S) results in a peak at [M + 2H − 74.01902]+•, and the Ile side-chain loss with an additional ammonia loss (C4H8 + NH3) at [M + 2H − 73.0891]+•. Although these interfering peaks could lead to erroneous identification of N-terminal isoAsp, they do not usually pose a problem if the analysis is performed on an instrument with sufficient resolving power. For example, differentiating the singly-charged N-terminal isoAsp signature fragment ion at m/z = 1000 from the interfering Met side-chain loss ion would require a mass resolving power of > 190,000, which is easily achievable on an FTICR mass spectrometer such as the one used here.
N-terminal acetylation
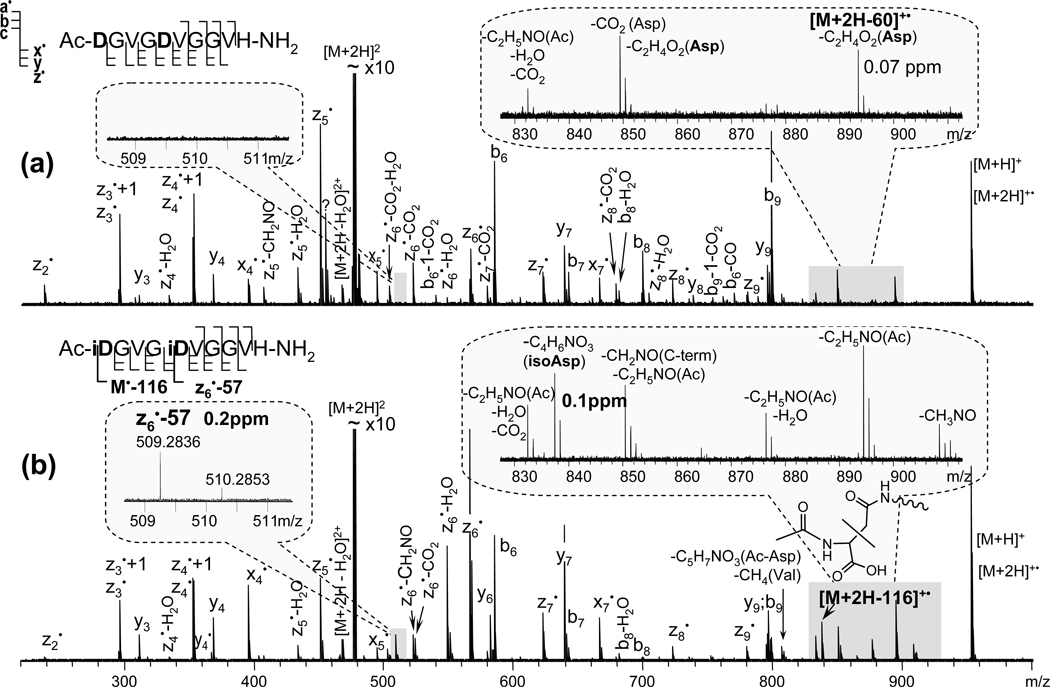
For experiments conducted on instruments with limited mass resolving power, such as ETD analysis done on an ion trap instrument, the peak interference problem could be circumvented by shifting the diagnostic peak out of the small molecule and side-chain loss region. This can be done by, for example, N-terminal acetylation. In this case, the C-terminal diagnostic fragment would be [M + 2H − (C2H5NO + C2HO2•)]+• = [M + 2H − 116.0348]+• (hereinafter indicated as [M + 2H − 116]+•), and the N-terminal diagnostic fragment would be C4H7NO3+• with m/z of 117.042. The effect of N-terminal acetylation was examined on the synthetic peptide Ac-DGVGDVGGVH-NH2 and its isoAsp-variant Ac-iDGVGiDVGGVH-NH2 using ECD. Specific diagnostic fragment peaks were observed for both isomers (Figure 3, insets): [M + 2H − 116]+• was present exclusively in the isoAsp-peptide spectrum, and the neutral loss of 60 (C2H4O2 = 60.0211) was only observed from the charge-reduced species of the Asp-peptide variant, indicated as [M + 2H − 60]+•. The peak at m/z = 894.4195 in the isoAsp-peptide spectrum (Figure 3, lower panel inset) is not related and corresponds to the neutral loss of 59 (C2H5NO = 59.0371) from the acetylated N-terminus. In addition, many of the fragments containing the Asp residue of the Asp-peptide also exhibit an accompanying loss of CO2 (Figure 3, upper panel); however, there are no such losses observed in the isoAsp-peptide ECD spectrum, except for the z6• − CO2. This ion was produced as a result of a secondary reaction following the z6• fragment ion formation via hydrogen rearrangement that only occurs when the Cα radical formed is adjacent to the isoAsp side chain. Similarly, a peak at m/z = 832 also has the CO2 loss, which could have been formed as a result of a secondary reaction upon the loss of an acetamide (C2H5NO).
Figure 3.
ECD spectra of the synthetic peptide variants with Asp (a) and isoAsp residues (b) amidated at the C-terminus and acetylated at the N-terminus. IsoAsp1 and isoAsp5 specific fragments, [M + 2H − 116]+• and z6• − 57, are observed only for the isoAsp-peptide variant. Asp side-chain losses are also indicated, (a) inset.
It should be noted that, if the N-terminally acetylated peptide contains a tryptophan (Trp) residue, the Trp side-chain loss of C8H6N• (116.05002) would interfere with the isoAsp diagnostic fragment peak [M + 2H − 116.0348]+• (to correctly identify such fragments at m/z ~ 1000, the minimum required resolving power is 65,000). In addition, no N-terminal isoAsp diagnostic fragment ion was observed in the N-terminally acetylated peptide, likely because of its small size and lack of a charge carrier. Therefore, other types of N-terminal elongation could be used for a more confident identification of the N-terminal isoAsp. Presumably, the use of larger tags may help to avoid undesired interference from the side-chain losses and for the detection of the N-terminal diagnostic ion.
Analysis of the N-terminal isoAsp by ETD
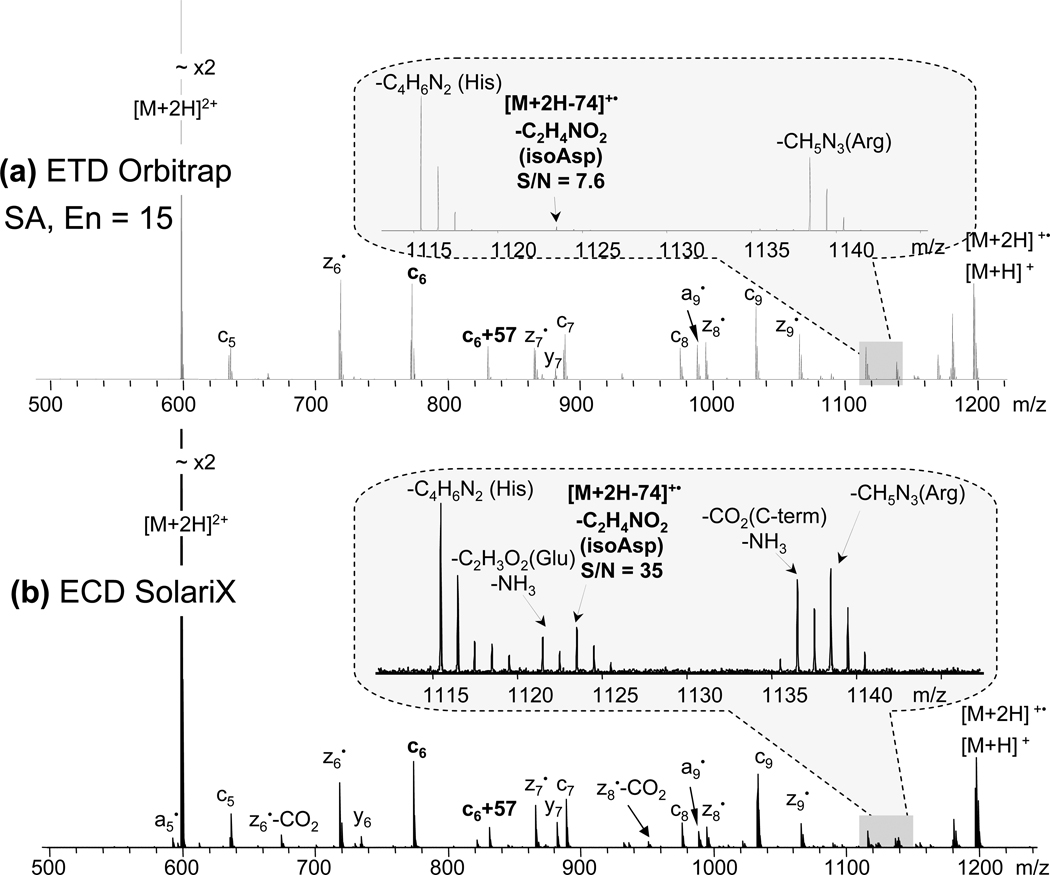
Peptides with N-terminal isoAsp were also analyzed using ETD. The N-terminal isoAsp diagnostic fragment peak [M + 2H − 74]+• was observed for all peptides, although in some cases, diagnostic peak detection was challenging as it exhibited low intensity. In particular, in ETD (LTQ-Orbitrap) of isoAsp-containing Aβ1–10 peptide, the peak corresponding to the diagnostic fragment ion was barely observable above the noise threshold level even with the supplemental activation (figure 4a, inset). In addition, some low-abundance peaks were barely detectable or absent in the ETD spectrum, e.g. a5•, and z7• − CO2. ECD provided 4.5 times larger S/N ratio for the diagnostic fragment peak (figure 4b, inset), possibly owing to the higher energy available in the ECD process than in the ETD process. Thus, the effect of additional energy input on diagnostic ion generation was further explored by means of supplemental activation.
Figure 4.
N-terminal isoAsp residue diagnostic fragment peak [M + 2H − 74]+• detection for the Aβ1–10 peptide variant (iDAEFRHiDSGY, fraction XI in the figure 3, top panel) by: (a) ETD on LTQ-Orbitrap, with supplemental activation energy parameter 15; and (b) ECD on SolariX. Insets show diagnostic peaks and their S/N ratios.
The effect of ion activation
Supplemental activation can improve fragment detection.39, 47, 48 Thus, the effect of supplemental ion activation (via SA, hotECD, and SD) on the diagnostic fragment peak intensity was investigated for all isoAsp-peptides studied. The S/N ratios and the relative intensities (Rel. Int.) of the diagnostic ion peaks are presented in Table 1. At first, the Rel. Int. was calculated, but it did not reflect the true picture for some peptides, because the absolute intensity of all fragments could change significantly with ion activation. Absolute S/N ratio seemed to be a better indicator than relative intensity, for example, with the Ang II ETD spectrum acquired on the amaZon instrument, application of SD resulted in a five-fold increase in the S/N ratio of the diagnostic fragment peak, while the Rel. Int. value did not change much (Table 1).
Table 1.
The effect of additional energy input on the diagnostic fragment formation. Relative intensity (Rel. Int.) was calculated as the absolute intensity of the diagnostic fragment divided by the sum of the absolute intensities of all c and z fragments.
| Peptide | Angiotensin II (iDRVYIHPF) |
Amyloid β (iDAEFRHiDSGY) |
Synthetic (Ac-iDGVGiDVGGVH-NH2) |
|||
|---|---|---|---|---|---|---|
| Signature ion | [M+2H−74]+• | [M+2H−74]+• | [M+2H−116]+• | |||
| m/z | 973.5259 | 1123.4807 | 837.4218 | |||
| Method | S/N | Rel. Int. | S/N | Rel. Int. | S/N | Rel. Int. |
| ETD Orbitrap no SA | 8.2 | 0.04 | 6.6 | 0.0022 | 316.3 | 0.0793 |
| ETD Orbitrap SA, En=5 | 40.5 | 0.053 | 6.4 | 0.0022 | 313.6 | 0.0803 |
| ETD Orbitrap SA, En=15 | 21.3 | 0.032 | 7.6 | 0.0014 | 293.9 | 0.0706 |
| ECD solariX | 104.6 | 0.029 | 35.0 | 0.0095 | 120.5 | 0.0549 |
| hotECD solariX 4.5V | 128.9 | 0.025 | 28.0 | 0.0096 | 291.3 | 0.0524 |
| ETD solariX | 145.8 | 0.059 | 4.0 | 0.0044 | 98.4 | 0.0828 |
| ETD amaZon no SD | 17.7 | 0.057 | 8.2 | 0.007 | 160.7 | 0.0786 |
| ETD amaZon SD | 92.2 | 0.052 | 17.3 | 0.0036 | 162.3 | 0.0818 |
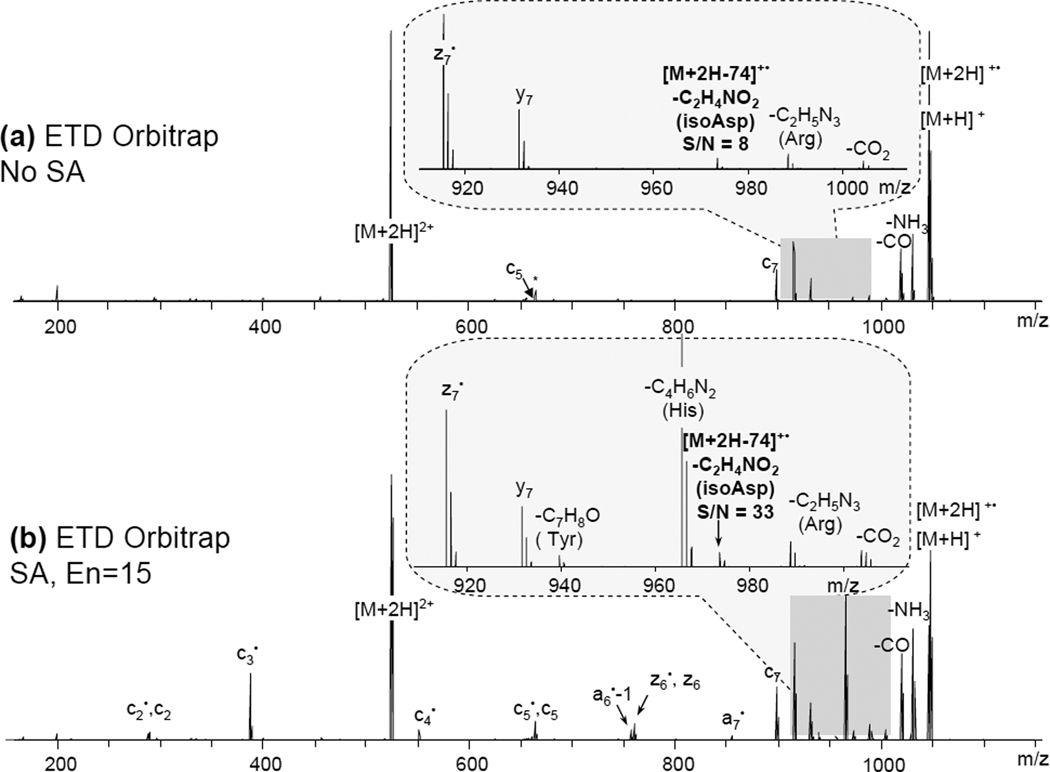
A direct comparison of the results acquired on different instruments is not possible because of the difference in multiple operating parameters, energetics of the reactions and the mass analyzers. When comparing spectra obtained using the same instrument, the results suggest that supplemental activation generally leads to higher S/N ratios for diagnostic ions. The effect of additional energy input on fragmentation depends on the peptide sequence, particularly, on the presence of basic residues and side-chain interactions. An example is shown on the ETD data acquired on an Orbitrap instrument with or without SA for the modified Ang II peptide (Figure 5). Similar to the ETD results acquired on an amaZon instrument mentioned earlier, the S/N ratio of the diagnostic ion became 4 times greater when SA was applied (Figure 5b).
Figure 5.
N-terminal isoAsp residue diagnostic fragment peak [M + 2H − 74]+• detection using ETD without (a) or with SA (b) for the modified Ang II peptide variant (iDRVYIHPF). Both spectra were acquired on Orbitrap. Insets show the S/N ratio of the [M + 2H − 74]+• peak and the appearance of additional side-chain losses upon SA (His and Tyr).
Conclusions
The N-terminal isoAsp residue was differentiated from Asp using ECD, hotECD, and ETD. Based on the detection of the specific fragment [M + 2H − 74]+• (isoAsp1), c6 + 57 (isoAsp7), and [M + 2H − 60]+• (Asp1, Asp7) it was possible to differentiate a mixture of 9 peptides containing isoAsp, Asp, and Asn residues, produced upon partial deamidation of the Aβ1–10 (N1N7) peptide and separated by a RP-HPLC system. This result illustrates that HPLC separation followed by ExD analysis can be a powerful method for isoAsp-containing peptide mixture differentiation. Although the detection of the N-terminal diagnostic fragment peak can be challenging, it is possible to substantially improve its detection by application of supplemental ion activation. In some cases, the S/N ratio of the signature fragment ion peak was increased by up to 6 times. Several interfering peaks may further complicate the analysis when Met or Ile amino acids are present. This can be overcome by the N-terminal elongation, with, for example, acetylation, which introduces a mass shift of the diagnostic peak to the smaller m/z region where fewer side-chain losses are available. However, if a Trp residue is present in the sequence, other types of elongation would be preferred. In addition, with a larger tag attached to the N-terminus, an N-terminal diagnostic peak may become detectable.
Supplementary Material
Figure S1. ECD spectra of chromatographic peaks I, IV–VII from the partially deamidated Aβ1–10 (N1N7) peptide mixture. Spectra for fractions II and III are not shown as they consist of unrelated peptides. The data is available upon request.
ACKNOWLEDGMENT
The authors acknowledge Eugene Moskovets and Alexander Cherkassky for helpful discussions. This work was supported by the NIH/NCRR-P41 RR10888, NIH/NIGMS-R01GM078293, and NIH/NCRR-S10 RR025082 grants. The Warwick University Chemistry Department and Warwick Centre for Analytical Science (EPSRC funded EP/F034210/1) are gratefully acknowledged.
REFERENCES
- 1.Clarke S. Int. J. Pept. Protein Res. 1987;30:808–821. doi: 10.1111/j.1399-3011.1987.tb03390.x. [DOI] [PubMed] [Google Scholar]
- 2.Geiger T, Clarke S. J. Biol. Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 3.Harris RJ, Kabakoff B, Macchi FD, Shen FJ, Kwong M, Andya JD, Shire SJ, Bjork N, Totpal K, Chen AB. J Chromatogr B. 2001;752:233–245. doi: 10.1016/s0378-4347(00)00548-x. [DOI] [PubMed] [Google Scholar]
- 4.Xiao G, Bondarenko PV, Jacob J, Chu GC, Chelius D. Anal. Chem. 2007;79:2714–2721. doi: 10.1021/ac0617870. [DOI] [PubMed] [Google Scholar]
- 5.Stephenson RC, Clarke S. J. Biol. Chem. 1989;264:6164–6170. [PubMed] [Google Scholar]
- 6.Robinson NE, Robinson AB. Proc. Natl. Acad. Sci. U. S. A. 2001;98:944–949. doi: 10.1073/pnas.98.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritz-Timme S, Collins MJ. Ageing Res. Rev. 2002;1:43–59. doi: 10.1016/s0047-6374(01)00363-3. [DOI] [PubMed] [Google Scholar]
- 8.Fujii N, Ishibashi Y, Satoh K, Fujino M, Harada K. Biochim. Biophys. Acta. 1994;1204:157–163. doi: 10.1016/0167-4838(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 9.Roher AE, Lowenson JD, Clarke S, Wolkow C, Wang R, Cotter RJ, Reardon IM, Zurcherneely HA, Heinrikson RL, Ball MJ, Greenberg BD. J. Biol. Chem. 1993;268:3072–3083. [PubMed] [Google Scholar]
- 10.Shimizu T, Matsuoka Y, Shirasawa T. Biol. Pharm. Bull. 2005;28:1590–1596. doi: 10.1248/bpb.28.1590. [DOI] [PubMed] [Google Scholar]
- 11.Noguchi S. Biopolymers. 2010;93:1003–1010. doi: 10.1002/bip.21514. [DOI] [PubMed] [Google Scholar]
- 12.Rehder DS, Chelius D, McAuley A, Dillon TM, Xiao G, Crouse-Zeineddini J, Vardanyan L, Perico N, Mukku V, Brems DN, Matsumura M, Bondarenko PV. Biochemistry. 2008;47:2518–2530. doi: 10.1021/bi7018223. [DOI] [PubMed] [Google Scholar]
- 13.Hambly DM, Banks DD, Scavezze JL, Siska CC, Gadgil HS. Anal. Chem. 2009;81:7454–7459. doi: 10.1021/ac901258g. [DOI] [PubMed] [Google Scholar]
- 14.Kameoka D, Ueda T, Imoto T. J. Biochem. 2003;134:129–135. doi: 10.1093/jb/mvg120. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Czupryn JM, Boyle PT, Jr, Amari J. Pharm. Res. 2002;19:1223–1231. doi: 10.1023/a:1019814713428. [DOI] [PubMed] [Google Scholar]
- 16.Lapko VN, Purkiss AG, Smith DL, Smith JB. Biochemistry. 2002;41:8638–8648. doi: 10.1021/bi015924t. [DOI] [PubMed] [Google Scholar]
- 17.Johnson BA, Aswad DW. Biochemistry. 1990;29:4373–4380. doi: 10.1021/bi00470a017. [DOI] [PubMed] [Google Scholar]
- 18.Reissner KJ, Aswad DW. Cell. Mol. Life Sci. 2003;60:1281–1295. doi: 10.1007/s00018-003-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Donato A, Ciardiello MA, de Nigris M, Piccoli R, Mazzarella L, D'Alessio G. J. Biol. Chem. 1993;268:4745–4751. [PubMed] [Google Scholar]
- 20.Aswad DW, Paranandi MV, Schurter BT. 3rd Symposium on the Analysis of Well Characterized Biotechnology Pharmaceuticals; Jan 07, 1999; Washington, D.C.. pp. 1129–1136. [Google Scholar]
- 21.Alfaro JF, Gillies LA, Sun HG, Dai SJ, Zang TZ, Klaene JJ, Kim BJ, Lowenson JD, Clarke SG, Karger BL, Zhou ZS. Anal. Chem. 2008;80:3882–3889. doi: 10.1021/ac800251q. [DOI] [PubMed] [Google Scholar]
- 22.Shin Y, Cho HS, Fukumoto H, Shimizu T, Shirasawa T, Greenberg SM, Rebeck GW. Acta Neuropathol. 2003;105:252–258. doi: 10.1007/s00401-002-0639-0. [DOI] [PubMed] [Google Scholar]
- 23.Ni W, Dai S, Karger BL, Zhou ZS. Anal. Chem. 2010;82:7485–7491. doi: 10.1021/ac101806e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terashima I, Koga A, Nagai H. Anal. Biochem. 2007;368:49–60. doi: 10.1016/j.ab.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Li XJ, Cournoyer JJ, Lin C, O'Cormora PB. J. Am. Soc. Mass Spectrom. 2008;19:855–864. doi: 10.1016/j.jasms.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Czupryn MJ. J. Pharm. Biomed. Anal. 2003;30:1479–1490. doi: 10.1016/s0731-7085(02)00479-x. [DOI] [PubMed] [Google Scholar]
- 27.De Boni S, Oberthur C, Hamburger M, Scriba GKE. J. Chromatogr. A. 2004;1022:95–102. doi: 10.1016/j.chroma.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Krokhin OV, Antonovici M, Ens W, Wilkins JA, Standing KG. Anal. Chem. 2006;78:6645–6650. doi: 10.1021/ac061017o. [DOI] [PubMed] [Google Scholar]
- 29.Winter D, Pipkorn R, Lehmann WD. J. Sep. Sci. 2009;32:1111–1119. doi: 10.1002/jssc.200800691. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann WD, Schlosser A, Erben G, Pipkorn R, Bossemeyer D, Kinzel V. Protein Sci. 2000;9:2260–2268. doi: 10.1110/ps.9.11.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez LJ, Shimizu T, Satomi Y, Betancourt L, Besada V, Padron G, Orlando R, Shirasawa T, Shimonishi Y, Takao T. Rapid Commun. Mass Spectrom. 2000;14:2092–2102. doi: 10.1002/1097-0231(20001130)14:22<2092::AID-RCM137>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 32.Castet S, Enjalbal C, Fulcrand P, Guichou JF, Martinez J, Aubagnac JL. Rapid Commun. Mass Spectrom. 1996;10:1934–1938. [Google Scholar]
- 33.Yamazaki Y, Fujii N, Sadakane Y. Anal. Chem. 2010;82:6384–6394. doi: 10.1021/ac100310x. [DOI] [PubMed] [Google Scholar]
- 34.Cournoyer JJ, Pittman JL, Ivleva VB, Fallows E, Waskell L, Costello CE, O'Connor PB. Protein Sci. 2005;14:452–463. doi: 10.1110/ps.041062905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cournoyer JJ, Lin C, O'Connor PB. Anal. Chem. 2006;78:1264–1271. doi: 10.1021/ac051691q. [DOI] [PubMed] [Google Scholar]
- 36.O'Connor PB, Cournoyer JJ, Pitteri SJ, Chrisman PA, McLuckey SA. J. Am. Soc. Mass Spectrom. 2006;17:15–19. doi: 10.1016/j.jasms.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Cournoyer JJ, Lin C, Bowman MJ, O'Connor PB. J. Am. Soc. Mass Spectrom. 2007;18:48–56. doi: 10.1016/j.jasms.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Sargaeva NP, Lin C, O'Connor PB. Anal. Chem. 2009;81:9778–9786. doi: 10.1021/ac901677t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan WYK, Chan TWD, O'Connor PB. J. Am. Soc. Mass Spectrom. 2010;21:1012–1015. doi: 10.1016/j.jasms.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee R, Adhikary L, Khedkar A, Iyer H. Rapid Commun. Mass Spectrom. 2010;24:879–884. doi: 10.1002/rcm.4464. [DOI] [PubMed] [Google Scholar]
- 41.Yang HQ, Fung EYM, Zubarev AR, Zubarev RA. J. Proteome Res. 2009;8:4615–4621. doi: 10.1021/pr900428m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreazza HJ, Wang T, Bagley CJ, Hoffmann P, Bowie JH. Rapid Commun. Mass Spectrom. 2009;23:1993–2002. doi: 10.1002/rcm.4107. [DOI] [PubMed] [Google Scholar]
- 43.Sargaeva NP, Goloborodko AA, Moskovets E, Gorshkov MV, O'Connor PB. Electrophoresis. doi: 10.1002/elps.201000507. in press. [DOI] [PubMed] [Google Scholar]
- 44.Falth M, Savitski MM, Nielsen ML, Kjeldsen F, Andren PE, Zubarev RA. Anal. Chem. 2008;80:8089–8094. doi: 10.1021/ac800944u. [DOI] [PubMed] [Google Scholar]
- 45.Robinson NE, Robinson ZW, Robinson BR, Robinson AL, Robinson JA, Robinson ML, Robinson AB. J. Pept. Res. 2004;63:426–436. doi: 10.1111/j.1399-3011.2004.00151.x. [DOI] [PubMed] [Google Scholar]
- 46.Robinson NE, Robinson AB. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horn DM, Ge Y, McLafferty FW. Anal. Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 48.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Anal. Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. ECD spectra of chromatographic peaks I, IV–VII from the partially deamidated Aβ1–10 (N1N7) peptide mixture. Spectra for fractions II and III are not shown as they consist of unrelated peptides. The data is available upon request.