Abstract
Filamentous fungi produce an impressive variety of secondary metabolites; many of them have important biological activities. The biosynthesis of these secondary metabolites is frequently induced by plant-derived external elicitors and appears to also be regulated by internal inducers, which may work in a way similar to that of bacterial autoinducers. The biosynthesis of penicillin in Penicillium chrysogenum is an excellent model for studying the molecular mechanisms of control of gene expression due to a good knowledge of the biochemistry and molecular genetics of β-lactam antibiotics and to the availability of its genome sequence and proteome. In this work, we first developed a plate bioassay that allows direct testing of inducers of penicillin biosynthesis using single colonies of P. chrysogenum. Using this bioassay, we have found an inducer substance in the conditioned culture broths of P. chrysogenum and Acremonium chrysogenum. No inducing effect was exerted by γ-butyrolactones, jasmonic acid, or the penicillin precursor δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine. The conditioned broth induced penicillin biosynthesis and transcription of the pcbAB, pcbC, and penDE genes when added at inoculation time, but its effect was smaller if added at 12 h and it had no effect when added at 24 h, as shown by Northern analysis and lacZ reporter studies. The inducer molecule was purified and identified by mass spectrometry (MS) and nuclear magnetic resonance (NMR) as 1,3-diaminopropane. Addition of pure 1,3-diaminopropane stimulated the production of penicillin by about 100% compared to results for the control cultures. Genes for the biosynthesis of 1,3-diaminopropane have been identified in the P. chrysogenum genome.
INTRODUCTION
Filamentous fungi, particularly ascomycetes and basidiomycetes, are well-known producers of hundreds of bioactive secondary metabolites (40). The recent sequencing of the genomes of Aspergillus niger (28), Aspergillus nidulans (12, 50), Aspergillus oryzae (25), Penicillium chrysogenum (49), and the basidiomycete Pleurotus ostreatus (G. Pisabarro, personal communication) has provided evidence about the presence in their genomes of numerous gene clusters encoding nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS), hybrid polyketide-nonribosomal peptide synthetases (HPS), isoprenoid synthetases, and other nonconventional antibiotic-synthesizing enzymes. Genome mining is a powerful tool for discovering new bioactive natural products (38).
Penicillin biosynthesis in P. chrysogenum is an excellent model for understanding how those complex molecules are synthesized and how the cell controls their production, because its biochemistry and molecular genetics are very well known (26). Penicillin is produced by several filamentous fungi, of which P. chrysogenum (26), A. nidulans (4), and Penicillium nalgiovense (20) have been studied in more detail. The three enzymes α-aminoadipyl-cysteinyl-valine synthetase (ACVS), isopenicillin N synthase (IPNS), and isopenicillin N acyltransferase (IAT) are encoded, respectively, by the pcbAB, pcbC, and penDE genes, which are linked together in the so-called pen cluster (10, 11, 48).
Expression of the secondary metabolite gene clusters in wild-type fungal strains in soil, plant tissues, or aquatic habitats is frequently very low (reflecting a low need for these metabolites in their natural role in the producer strains) and is modulated in response to different nutritional or environmental stimuli (20, 38). In some cases many of the secondary metabolite genes remain silent, although it is likely that they may be expressed under still-unknown conditions (38).
In plant-pathogenic fungi, production of secondary metabolites is elicited by plant materials, named elicitors. The plant reacts to the fungal infection by producing antifungal plant metabolites that act as defenses against the fungal attack. Some of the plant materials (e.g., alginates or oligomannuronate) have been used to stimulate penicillin biosynthesis (2, 13).
Autoinducers are well known regulators of secondary metabolism and differentiation in bacteria (6, 15, 33, 53), but they are largely unknown in fungi (34, 35). There are different types of autoregulatory molecules (32, 53). They include homoserine lactones, γ-butyrolactones, modified nucleotides (such as the B factor), modified peptides (17), and other small molecules, such as 2,3-diamino-2,3-bis(hydroxymethyl)-1,4-butanediol (PI factor) (32). The possible presence of autoregulatory molecules in fungi has received little attention, although they likely play very important roles in the complex differentiation processes of these organisms (34, 35).
The presence of nonribosomal peptides in the culture of the penicillin or cephalosporin producers P. chrysogenum, Acremonium chrysogenum, and Paecilomyces persicinus (8, 23) that are secreted into the culture broth (24) led us to study if these peptides or another secreted small molecule serves to induce penicillin biosynthesis.
In this work, we describe the presence of an autoinducer molecule in the culture broths of both P. chrysogenum and A. chrysogenum in defined medium. The results described in this article show that the penicillin inducer molecule synthesized by those fungi is 1,3-diaminopropane.
MATERIALS AND METHODS
Strains and culture conditions.
P. chrysogenum NRRL 1951 (wild-type strain), P. chrysogenum Wisconsin 54-1255 (reference strain for the genome sequencing project, here referred as Wis 54-1255), and P. chrysogenum npe10 (Δpen), which is derived from the Wis 54-1255 strain and lacks the penicillin gene cluster (10), were used in this work. They were grown on solid Power medium (5) for 7 days at 28°C. P. chrysogenum NRRL 1951 and Wis 54-1255 were used for testing the presence of autoinducers of penicillin biosynthesis by bioassay (see below), whereas P. chrysogenum Wis 54-1255 and P. chrysogenum npe10 were used to obtain conditioned culture broths. Conidia from these two strains grown on one petri dish were collected and inoculated into a flask with 100 ml of defined DP medium (5) without phenylacetate. After incubation at 25°C for 20 h in an orbital shaker (250 rpm), a 10-ml aliquot was inoculated in 500-ml flasks containing 100 ml DP medium with 1 mg/ml phenylacetate (for benzylpenicillin biosynthesis).
A. chrysogenum N2, a cephalosporin nonproducer mutant (42), which is deficient in isopenicillin N synthase (30) due to a mutation in the structural pcbC gene and therefore accumulates the tripeptide ACV (31), was used to obtain conditioned culture broths. This strain was grown in plates of LPE medium (per liter: glucose, 1 g; yeast extract, 2 g; NaCl, 1.5 g; CaCl2, 10 g; 2% [wt/vol] agar, pH 6.8) for 7 days at 28°C. Spores and mycelium fragments collected from six plates of LPE culture medium were inoculated in 100 ml of seed medium (41) in 500-ml shake flasks and incubated at 25°C for 48 h in an orbital incubator at 250 rpm. Ten milliliters of this seed culture was used to inoculate 100 ml of defined cephalosporin production medium (29) in triple-baffled flasks (500 ml) and incubated at 25°C in a rotary shaker (250 rpm). In order to ensure enough culture medium for the purification of the inducer molecule, fermentations with A. chrysogenum N2 were run (15 liters) at 25°C and 300 rpm in a Braun Biostat C20 fermentor using the Shen medium (41).
The yeast Candida utilis (syn. Pichia jadinii) CETCT 1061 was also used to obtain conditioned culture broths. This strain was grown in TSA (tryptone soya broth [TSB] agar) plates for 1 day at 28°C. Cells were collected from three plates and inoculated in 100 ml of YED medium (per liter: 10 g yeast extract, 10 g glucose, 4 g (NH4)2SO4, pH 7.0) in 500 ml shake flasks.
Conditioned culture broths of P. chrysogenum Wis 54-1255, P. chrysogenum npe10, and A. chrysogenum N2 were obtained after 72 h of growth, whereas conditioned culture broths of C. utilis were obtained after 48 h of growth (due to the higher specific growth rate of this yeast). Cultures were filtered through nylon filters (Nytal Maissa, Barcelona, Spain) and then centrifuged at 4,000 × g. The clear supernatants were used for the inducer assay directly or concentrated 2- to 10-fold.
Qualitative solid test for autoinducers.
Two microliters of a solution containing 1 × 106 spores/ml of P. chryogenum NRRL 1951 or Wis 54-1255 was inoculated into defined Czapek Dox medium. Plates were incubated at 28°C for 24 h. After incubation, a 5-mm-diameter well was excavated adjacent to the edge of individual colonies. Different compounds and solutions were added (100 μl) inside the well (milliQ water was used as a control), and after 24 h at 28°C, an overlay containing 1% TSA (Difco) with spores of Bacillus subtilis ATCC 6633 (100 μl of spore suspension per 100 ml of 1% TSA) was added. Plates were incubated overnight at 30°C. Induction of antibiotic production gave rise to inhibition halos of the B. subtilis lawn.
Quantitation of penicillin production.
Production of total penicillins was quantified by bioassay using Micrococcus luteus as a test microorganism. It was grown at an optical density at 600 nm (OD600) of 2 in TSB (Difco) and inoculated into TSA (Difco) medium at a final OD600 of 0.01. One hundred microliters of the filtrates obtained from P. chrysogenum cultures grown on liquid DP medium (5) were added to 5-mm-diameter wells dug in the TSA plates. After 2 h at 4°C, plates were incubated for 15 h at 30°C. Benzylpenicillin production was assessed by high-performance liquid chromatography (HPLC) as previously described (14).
Northern blot.
Total RNA from P. chrysogenum was isolated from mycelia grown in DP medium. Total RNA isolation and Northern analysis were carried out as described previously (46). Three different P. chrysogenum probes were used: the 996-bp pcbC gene, the 1,074-bp cDNA of the penDE gene, and the 0.8-kb NcoI-KpnI internal fragment of the β-actin gene. Quantitation of the radioactivity intensity (cpm) was performed with a phosphorimager scanner (Instantimager; Packard Instrument Company).
Cell extracts and β-galactosidase assays.
Protein extracts were obtained from mycelia grown in DP medium, and β-galactosidase activity was determined as previously described (18).
Extraction studies of the inducer molecule.
Cultures obtained from fermentations in a Braun Biostat C20 stirred tank fermentor (15 liters) were filtered through nylon filters (Nytal), concentrated under vacuum to about one-third of the original volume, and deproteinated by acidification to pH 2.0 with 1 M HCl in an ice bath (4°C) for 8 h. The proteinaceous precipitate was removed by centrifugation at 8,000 rpm (5,900 × g) for 90 min. To determine the hydrophilic or hydrophobic (organic solvent-soluble) nature of the inducer molecule(s), the deproteinated broth was extracted with ethylacetate (3 volumes of ethylacetate per volume of concentrated deproteinated broth). The extraction was repeated three times. The organic or aqueous phase was concentrated to dryness under vacuum and redissolved in either methanol (the residue of organic phase) or milliQ water (the residue of the aqueous phase). Both fractions were tested for inducer activity and analyzed by HPLC.
Chromatographic methods.
The resin for the cationic exchange chromatography column was hydrated with 100 mM sodium acetate buffer (pH = 3.0). In the case of the resin of the anionic exchange chromatography column, the latter was embedded in 100 mM Tris-HCl (pH = 8.0). The resin in the column was equilibrated by adding 10 volumes of the corresponding buffer at a concentration of 10 mM.
For the hydrophobic interaction chromatography (phenyl-Sepharose fast flow), the resin was hydrated using milliQ water for 12 to 14 h at 4°C. After the resin was packaged in the column, it was equilibrated adding 10 volumes of milliQ water.
For gel filtration chromatography, Sephadex G10 resin was hydrated with milliQ water for 12 to 24 h at room temperature, and the packaged column was equilibrated with 10 volumes of 0.05 M sodium acetate.
HPLC analysis.
Samples obtained after purification through ion exchange and gel filtration chromatographies were derivatized with fluorenylmethyloxycarbonyl chloride (FMOC-Cl) (Sigma-Aldrich) as previously described (14). Samples were lyophilized and resuspended in 1 ml of milliQ water. Separation of the inducer was performed in a LiChrospher 100 RP-18 (4 by 250 mm), 5-μm (Merck) column using a gradient elution consisting of acetonitrile and sodium acetate buffer. The mobile phase was prepared as follows: solvent A, sodium acetate buffer, 50 mM, pH 4.2; solvent B, acetonitrile. The injection volume was 10 μl, with a flow rate of 1 ml/min. The elution gradient was as follows: 20% B → 25% B over 7 min; 25% B → 20% B over 8 min; 20% B→ 50% B for 15 min; 50% B → 70% B for 3 min; 70% B, linear for 5 min; 70% B → 20% B for 2 min; 20% B, linear for 6 min. Detection was performed at 254 nm.
Polyamine detection was achieved through derivatization with benzoyl chloride as follows. Samples (500 μl) were treated with 2 N NaOH and 10 μl of benzoyl chloride (Sigma-Aldrich). After vigorous vortexing for 10 s, the samples were incubated for 20 min at room temperature. Next, 2 ml of a saturated solution of NaCl was added, followed by the addition of 2 ml of diethyl ether and gentle mixing. Samples were centrifuged at 4,000 × g for 15 min. After centrifugation, 1 ml of the organic fraction was extracted and dried using a speed-vac machine. Samples were resuspended in HPLC-grade methanol. The separation was performed in a Teknokroma C18 (4.6 by 150 mm), 3-μm (Sea Mediterranea) column using a gradient elution consisting of milliQ water (solvent A) and methanol (solvent B). The injection volume was 10 μl, with a flow rate of 1 ml/min. The elution gradient was as follows: 60% B, linear for 7 min; 60 → 80% B for 4 min; 80% B → 100% B for 4 min; 100% B, linear for 4 min; 100% B → 60% B for 1 min; 60% B, linear for 4 min. Detection was performed at 254 nm.
RESULTS
Development of a bioassay for testing autoinducers of penicillin biosynthesis.
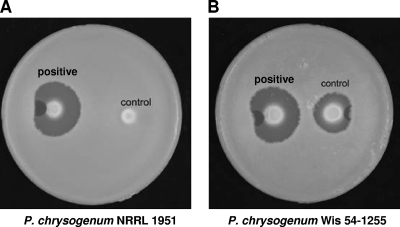
Autoinducers are usually growth-associated compounds synthesized by the producer strain that are secreted during the late growth phase into the so-called “conditioned broth” (meaning a broth that contains autoinducers). For routine testing of autoinducer molecules occurring in the conditioned broth of cultures of P. chrysogenum, A. chrysogenum, or other fungi, a qualitative solid test was developed (see Materials and Methods), in which the conditioned broth and compounds to be tested were added to a 5-mm-diameter well excavated in agar plates, adjacent to the edge of individual colonies of P. chrysogenum Wis 54-1255 or P. chrysogenum NRRL 1951 growing in defined Czapek Dox medium. The production of penicillin was visualized with an overlay of B. subtilis ATCC 6633 culture that formed a lawn. Under these conditions, colonies of the wild-type P. chrysogenum NRRL 1951 formed no detectable (or barely visible) inhibition halos (Fig. 1A), whereas colonies of P. chrysogenum Wis 54-1255 produced small inhibition halos (uninduced control strain) that increased in diameter in response to the inducer (Fig. 1B). When NRRL 1951 was used in the tests, it required longer incubation times in the presence of the inducer (48 h at 28°C instead of 24 h) to observe the induction effect.
Fig. 1.
Bioassays developed for testing autoinducers of penicillin biosynthesis. Individual colonies of wild-type P. chrysogenum NRRL 1951 (A) or the improved strain P. chrysogenum Wis 54-1255 (B) were grown in defined Czapek Dox medium agar plates. The conditioned broth containing the putative inducer was added inside a 5-mm well excavated in the agar adjacent to the edge of each colony. Note that there is a convex or plain distortion in the inhibition zone that corresponds to the place where the well was made in the agar, which prevents correct diffusion of the penicillin from the producer colony.
This test was also used to assess the possible inducing effect of homoserine lactone, butyrolactones (Sigma-Aldrich), and jasmonic acid (a plant secondary metabolite inducer) at concentrations in the range of 100 nM to 10 mM. As shown in the Fig. S1 in the supplemental material, none of those compounds induced penicillin biosynthesis.
We also assayed the possible inducing capability of the ACV tripeptide. As shown in Fig. S1 in the supplemental material, neither the monomeric or dimeric ACV nor the constituent amino acid cysteine or glycine was able to induce penicillin biosynthesis under these conditions. Similar negative results were obtained with each of the 20 proteinogenic amino acids (data not shown).
Conditioned culture broths of P. chrysogenum and A. chrysogenum contain a penicillin inducer.
Conditioned broths of P. chrysogenum Wis 54-1255 (which is expected to have a normal inducer level) and P. chrysogenum npe10 (which might lack the inducer), a Wis 54-1255 derivative that completely lacks the pen gene cluster region, were prepared as indicated in Materials and Methods. Conditioned broths of A. chrysogenum N2, a mutant that accumulates and secretes large amounts of the ACV intermediate and other peptides, were also tested.
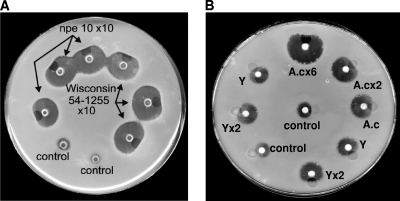
As shown in Fig. 2A, both the P. chrysogenum Wis 54-1255 and penicillin nonproducer mutant npe10 culture broths contained a putative inducer of penicillin biosynthesis; i.e., the formation of this putative inducer was not encoded by the 105-kb DNA region containing the pen cluster that is deleted in the npe10 mutant.
Fig. 2.
Bioassays of induction of penicillin biosynthesis by colonies of P. chrysogenum Wis 54-1255 using conditioned culture broths. (A) Conditioned culture broths of P. chrysogenum Wis 54-1255 and the penicillin nonproducer mutant npe10 in DP medium (72 h) concentrated 10 times were used for the bioassay. (B) Conditioned culture broths of A. chrysogenum N2 (A.c) in DP medium (72 h) and C. utilis (Y) in YED medium (48 h) were concentrated 2 and 6 times (for A. chrysogenum) or 2 times (for C. utilis) or used directly without any concentration step. MilliQ water was added as a control.
A similar induction level was observed using the conditioned culture broths of A. chrysogenum N2 (Fig. 2B); this inducing effect was not due to the tripeptide ACV, since, as shown in Fig. S1 in the supplemental material, ACV does not act as an inducer. Some induction activity was also observed in the conditioned broths of the yeast C. utilis in YED medium. Control assays with the culture medium without inoculation did not show any inducing effect.
Effect of conditioned broths on production of penicillin in liquid cultures of P. chrysogenum Wis 54-1255.
The effect of the conditioned broth of either P. chrysogenum Wis 54-1255 or A. chrysogenum N2 on penicillin production by P. chrysogenum was tested by adding it to the cultures at inoculation time (10 ml of conditioned broth plus 90 ml of DP culture medium).
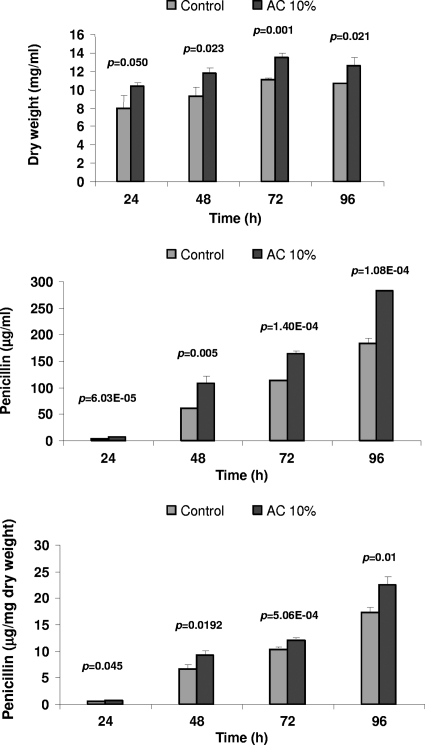
As shown in Fig. 3, the N2 conditioned broth when added at inoculation time stimulated the biomass formation of P. chrysogenum Wis 54-1255 by about 20% and increased significantly the volumetric (up to 70%) and specific penicillin production (per mg of dry weight) up to 33%. Similar results were obtained with the conditioned broths of P. chrysogenum npe10 and A. chrysogenum N2. Since the N2 strain is known to accumulate several extracellular peptides (42), more emphasis was dedicated to the characterization of the inducer molecule in the conditioned broth of the N2 strain.
Fig. 3.
Effect of the addition of conditioned broths on growth and penicillin production in liquid cultures of P. chrysogenum Wis 54-1255. Conditioned broths of A. chrysogenum N2 (AC) were added (10%; not concentrated) at inoculation time to cultures of P. chrysogenum Wis 54-1255. Dry weight and penicillin production were determined at different time points. MilliQ water was added at the same percentage (10%) as for the control. Data were subjected to one-way analysis if variance (ANOVA), and the P value is indicated. Similar results were obtained when conditioned broths obtained from P. chrysogenum npe10 were used (data not shown).
Conditioned broth induced expression of the pcbC and penDE penicillin biosynthesis genes.
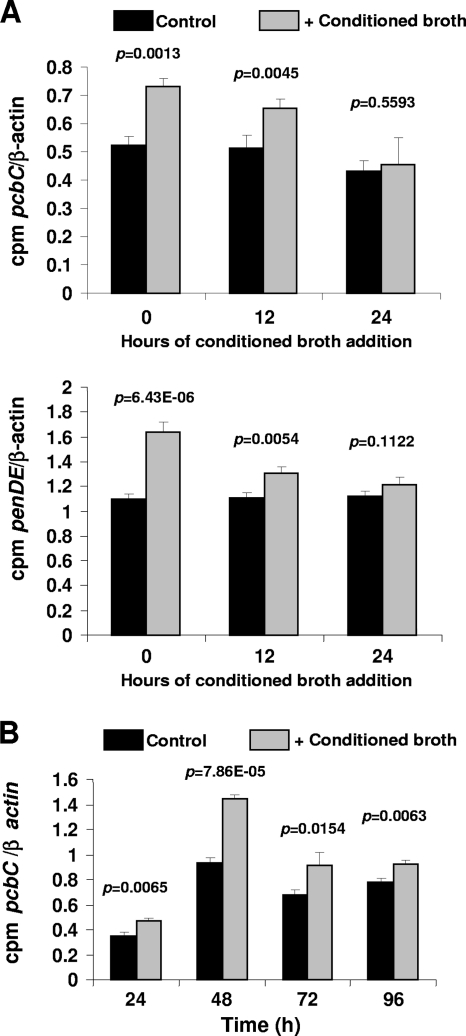
In order to assess if the effect of the conditioned culture broth was exerted at the transcription level, the conditioned broth was added to cultures at inoculation time (0 h), at 12 h, or at 24 h, and the expression of the pcbC and penDE genes was tested by Northern hybridization with radioactively labeled probes internal to the pcbC and penDE genes. RNAs were collected at 48 h. Results of this analysis confirmed that expression of the pcbC gene was clearly stimulated when the conditioned broth was added at 0 or 12 h, and the induction effect was clearly lower when added at 24 h (Fig. 4A; see also Fig. S1 in the supplemental material). Similarly, expression of the penDE gene was higher when the conditioned broth was added at 0 h or 12 h, and the effect was lower when it was added at 24 h (Fig. 4A; see also Fig. S2). These results indicate that the inducer has a greater effect when added before the secondary metabolite mRNAs are formed. This observation is consistent with the behavior of autoregulators in bacteria (15, 32).
Fig. 4.
Expression of the pcbC and penDE genes after the addition of conditioned broth. (A) Expression of the pcbC and penDE penicillin biosynthetic genes. The conditioned broth (10%) was added (+) to cultures at inoculation time (0 h) or 12 h or 24 h after the inoculation time. MilliQ water was added at the same percentage (10%) as for the control. RNA was extracted at 48 h. The transcripts from the β-actin gene were used as a control for the total RNA amount (see Fig. S2A in the supplemental material). Northern blots were hybridized with radioactively labeled pcbC, penDE, or β-actin probe and detected with a phosphorimager (Packard). The transcript levels were normalized, comparing the cpm provided by each mRNA to the β-actin mRNA radioactivity signal. (B) Quantitation of the radioactivity signal (cpm) provided in the Northern blot analysis shown in Fig. S2B. The transcript levels were normalized, comparing the cpm provided by each mRNA to the β-actin mRNA radioactivity signal. Data from panels A and B were subjected to one-way ANOVA, and the P value is indicated.
In order to confirm the effect of the inducer on the levels of the pcbC gene transcript during the growth phase, the conditioned broth was added at inoculation time (0 h), and RNA was extracted from cells taken at 24 h (the pcbC gene is a relatively early-expressed gene), 48 h, 72 h, and 96 h. Results of the Northern analysis (Fig. 4B; see also Fig. S2 in the supplemental material) showed that, indeed, the N2 conditioned broth already increased pcbC gene expression at 24 h, and the inducing effect was very clear in RNA samples of 48 and 72 h (up to a 47% increase compared to results for the control without conditioned broth).
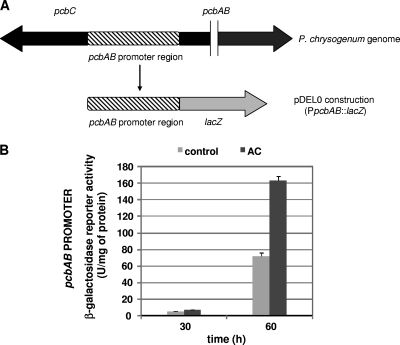
Induction effect of conditioned broth on expression of the pcbAB promoter coupled to the reporter lacZ gene.
Northern analysis of the very large pcbAB transcript (11.5 kb) always resulted in a smeared hybridization due to degradation of this large RNA during extraction and electrophoresis (7). Therefore, an alternative strategy was used to quantify the effect of the conditioned broth on pcbAB transcription. Expression of the pcbAB gene was quantified using the pDEL0 transformant, a derivative of P. chrysogenum npe10 in which the entire pcbAB promoter is coupled to the lacZ gene (Fig. 5A) and the construction is integrated as a simple copy in the pyrG locus of P. chrysogenum npe10 (18). Quantification of β-galactosidase activity showed that this activity was clearly higher at either 30 or 60 h of cultivation in the conditioned broth-supplemented cultures (at inoculation time) (Fig. 5B). In conclusion, our results indicate that the conditioned broth of strain N2 increases the expression of the three penicillin biosynthetic pathway genes pcbAB, pcbC, and penDE, particularly when added at inoculation time.
Fig. 5.
Effect of the conditioned broth on expression of the pcbAB promoter coupled to the reporter lacZ gene. (A) Map of the pDELO construction, containing the promoter of the pcbAB gene fused to the lacZ gene. (B) Quantification of the β-galactoxidase activity in control and conditioned broth-supplemented cultures (AC) of P. chrysogenum Wis 54-1255. The conditioned broth was added (10%) at inoculation time (0 h) to the pDEL0 strain. MilliQ water was added at the same percentage (10%) as for the control. Samples were isolated at 30 h and 60 h of culture, and β-galactosidase activity was measured using protein extracts obtained from those samples. Data represent the means and standard deviations from three independent experiments.
Concentration and purification of the inducer through ion exchangers, hydrophobic interaction chromatography, gel filtration chromatography, and HPLC.
The inducer molecule was purified successive to its activity by the agar penicillin induction assay. For this purpose, fermentations were run (15 liters) in a Braun Biostat C20 stirred tank fermentor using Shen medium (41). Extraction of the inducer molecule was carried out as indicated in Materials and Methods. The results showed that the inducer activity was present only in the aqueous phase but not in the organic (ethylacetate) extract.
Since the inducer molecule was water soluble and could not be extracted in the organic phase, its purification was carried out using strong cationic (Dowex-50-X8-400) or anionic (Dowex 1X8-400) ion exchangers. After several tests with both ion exchangers, the best purification was achieved by filtration through the cationic Dowex-50-X8-400 column in ammonium acetate buffer (pH 3.0). The inducer was eluted with a NaCl gradient (up to 0.5 M NaCl). A total of 120 3-ml fractions were collected, frozen at −80°C, and lyophilized. Each lyophilized fraction was resuspended in 400 μl of milliQ water, and sets of three consecutive fractions were pooled (numbered 1 to 40) and used for bioassays and HPLC analysis. As shown in Fig. S3 in the supplemental material, the activity was present in fractions 33 to 50 (mixtures 11 to 16), while there was no activity in the rest of the fractions.
Since the HPLC assay of the active fractions revealed the presence of several compounds, the preparation (mixture of the active fractions) was cleaned by filtration through a phenyl-Sepharose column in which the active inducer was not retained (see Fig. S3 in the supplemental material). Finally, the active fractions were subjected to chromatography in a Sephadex G-10 (GE Healthcare) column that allowed fractionation according to the molecular weights of the compounds. A total of 120 3-ml fractions were collected and lyophilized. Each lyophilized fraction was resuspended in 300 μl of milliQ water, and its inducer activity was analyzed by bioassay (see Fig. S4); the activity eluted in a clear peak (fractions f-31 to f-36). These fractions were mixed together in a single pool and analyzed by HPLC.
In HPLC analysis, five peaks were clearly separated (see Fig. S4 in the supplemental material), collected, and assayed in the penicillin inducing test. Only the product of peak 1 (fraction 1), with a retention time of 7 min, showed penicillin induction activity (see Fig. S4). HPLC purification was repeated several times until about 5 mg of pure compound was obtained.
MS and NMR spectrometry identified the inducer as 1,3-diaminopropane.
The structural elucidation of the compound in peak 1 was carried out by mass spectrometry (MS) and nuclear magnetic resonance (NMR) using a combination of one-dimensional NMR methods (1H NMR, 13C NMR, and distortionless enhancement by polarization transfer (DEPT) and two-dimensional shift-correlated NMR techniques (1H, 1H correlated spectroscopy [COSY], and heteronuclear multiple quantum correlation-heteronuclear single quantum correlation [HMQC-HSQC]), for the complete 1H and 13C signal assignments (Table 1). NMR was recorded in CDCl3 at room temperature in a Bruker WM 500 spectrometer (500 MHz for 1H NMR and 125 MHz for 13C NMR).
Table 1.
1H- and 13C-NMR chemical shift assignments of 1,3-diaminopropane in CDCl3
| Position no. | Group determined by DEPT | δ 13C (ppm) | δ 1H (ppm) |
|---|---|---|---|
| 1, 3 | CH2 | 39.08 | 2.34 (t, J = 6.9 Hz, 4H) |
| 2 | CH2 | 36.59 | 1.17 (m, 2H) |
Mass spectrometry showed an ion peak at m/z 75.1 [M + H]+, which revealed that its molecular weight was 74.0. Combined results of the MS and the 13C NMR data indicated that the molecular formula was C3H10N2.
The analysis of the 1H NMR spectrum showed two signals at δ 1.17 ppm and δ 2.34 ppm, which were assigned to methylene groups (CH2). This was confirmed by means of a DEPT study. Assignment of the carbon atoms of these methylene groups was carried out by using an HMQC-HSQC spectrum, which showed a correlation peak via 1JH,C with the methylene protons. The COSY experiment showed connectivities between the methylene protons at δ 1.17 ppm and the methylene protons at δ 2.34 ppm. Using the chemical shift values, it was fairly easy to establish the signal at δH-1,3 2.34 ppm (t, J = 6.9 Hz), δC-1,3 39.08 ppm, and the signals at δH-2 1.17 ppm (m), δC-2 36.59 ppm. This compound was identified as 1,3-diaminopropane, a low-molecular-weight diamine.
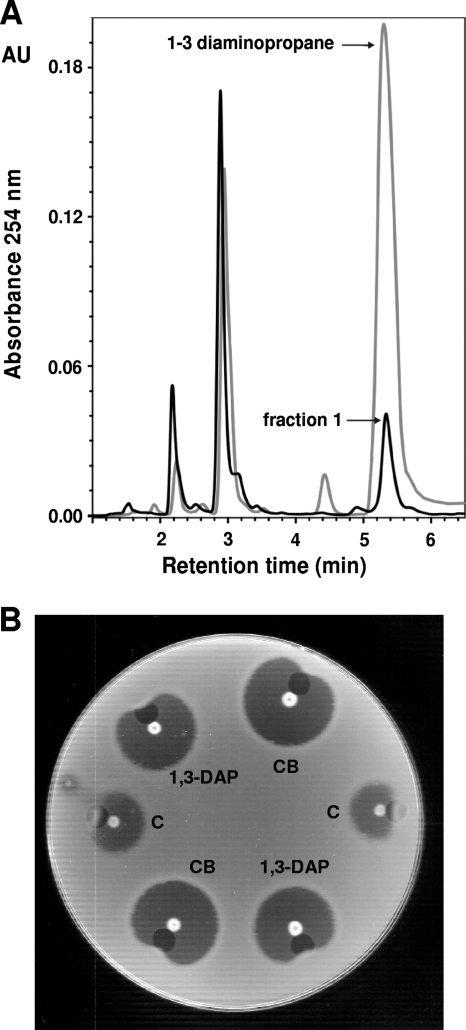
The peak of inducing activity derivatized with benzoyl chloride coeluted in HPLC with that of authentic 1,3-diaminopropane (Sigma-Aldrich) derivatized with the same reagent (Fig. 6A). The diaminopropane peak was also identified by analytical HPLC as the inducer of penicillin biosynthesis in the conditioned broth of P. chrysogenum Wis 54-1255 and P. chrysogenum npe10 (data not shown); i.e., the inducer molecule was the same in the broths of A. chrysogenum and P. chrysogenum. Quantitation of the diaminopropano peak indicated that the concentration of this compound in the conditioned broths (obtained after 72 h of growth) of these fungi ranged from 4 mM to 11 mM.
Fig. 6.
(A) HPLC analysis of the inducing fraction and 1,3-diaminopropane. Chromatogram showing the coelution of the positive fraction (number 1) with a sample of pure authentic 1,3-diaminopropane. Samples were derivatized with benzoyl chloride and detected at 254 nm, following the protocol described in Materials and Methods. Those peaks that occur both in fraction 1 and in the standard (until 3 min) correspond to free benzoyl chloride (excess of derivatizing agent). (B) Solid test bioassay carried out with 10 mM 1,3-diaminopropane and conditioned broths of A. chrysogenum (CB). The bioassay was performed in the same way as that shown in Fig. 1. Colonies of P. chrysogenum Wis 54-1255 were used, and milliQ water was added as a control.
Pure 1,3-diaminopropane was tested by qualitative solid bioassay (as indicated before) in parallel with the conditioned broths of A. chrysogenum. As shown in Fig. 6B, the inhibition halo generated by 1,3-diaminopropane (10 mM) was larger than that provided by the control, indicating that 1,3-diaminopropane exerted a positive effect on penicillin production This halo, however, was smaller than the one provided by the conditioned broths, suggesting that the inducer effect of 1,3-diaminopropane could be favored by different molecules that are present in the conditioned broth.
Exogenous 1,3-diaminopropane increases production of penicillin.
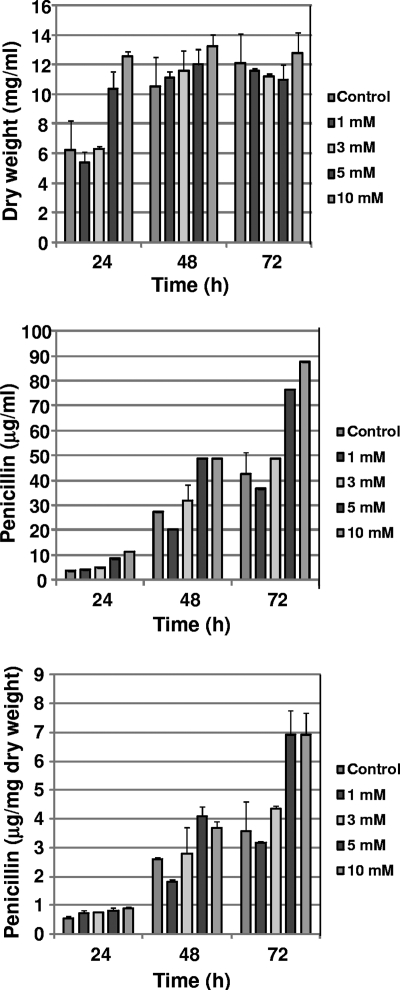
After the inducer molecule was identified, it was important to confirm that this compound stimulated the production of benzylpenicillin in liquid cultures. For this purpose, cultures (three biological replicates with two technical replicates each) were performed in defined DP medium supplemented with increasing concentrations of 1,3-diaminopropane. Results of the fermentation experiments (Fig. 7) showed that the maximal biomass of those cultures supplemented with either 5 or 10 mM 1,3-diaminopropane was reached at 24 h. At 48 h and 72 h, no significant differences were observed in the dry weight. Volumetric and specific penicillin titers (Fig. 7) significantly increased (around 100% compared to those of the control cultures) in those cultures supplemented with either 5 or 10 mM 1,3-diaminopropane, an effect which was specially remarkable after 72 h. These results confirmed the positive effect of 1,3-diaminopropane on penicillin production.
Fig. 7.
Effect of the addition of pure 1,3-diaminopropane on growth and penicillin production in liquid cultures of P. chrysogenum. Commercial 1,3-diaminopropane (Sigma-Aldrich) was added at different concentrations at inoculation time (0 h) to cultures of P. chrysogenum Wis 54-1255 grown in defined medium. Dry weight and penicillin production were estimated at different time points. The same volume of MilliQ water was added to the control cultures.
Effect of addition of putrescine, spermidine, and spermine on benzylpenicillin production.
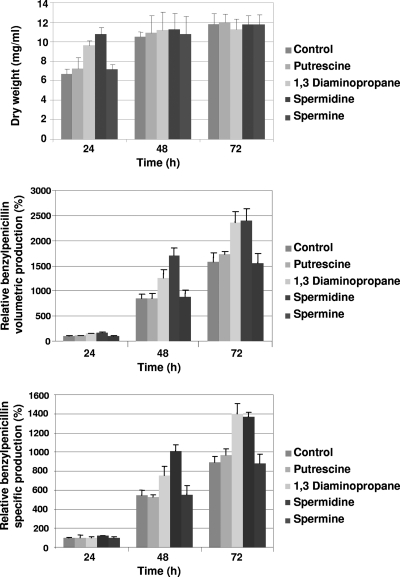
Since the inducer molecule 1,3-diaminopropane is a diamine, we decided to test the effect of other polyamines on penicillin production. With this aim, cultures (three biological replicates with two technical replicates each) were performed in defined DP medium supplemented with 1,3-diaminopropane and the polyamines putrescine, spermidine, and spermine (at a concentration of 5 mM each). Results of the fermentation experiments (Fig. 8) revealed that the culture achieved maximal biomass production at 24 h when 1,3-diaminopropane or spermidine was added. At 48 h and 72 h, no significant differences were observed in the dry weight. Measurement of benzylpenicillin by HPLC indicated that volumetric and specific titers increased around 60% (Fig. 8), especially at 48 h and 72 h, when 1,3-diaminopropane or spermidine was added. The addition of putrescine or spermine did not significantly modify benzylpenicillin production (see Discussion).
Fig. 8.
Effect of the addition of different polyamines on growth and penicillin production in liquid cultures of P. chrysogenum. Cultures of P. chrysogenum Wis 54-1255 grown in defined medium were supplemented at inoculation time (0 h) with either 1,3-diaminopropane, putrescine, spermidine, or spermine (5 mM each). The same volume of MilliQ water was added to the control cultures. Samples were taken at 24 h, 48 h, and 72 h. Benzylpenicillin was assessed by HPLC. Relative benzylpenicillin production (%) for each time point was calculated, setting the production that corresponds to the Wis 54-1255 strain at 24 h as 100%.
DISCUSSION
Autoinducers are very important signals that control several biochemical processes in the producer strains, frequently through “quorum-sensing” mechanisms. There are different chemical classes of autoinducers in microorganisms and plants (53). In several Gram-negative bacteria, homoserine lactones control bioluminescence, biofilm formation, pathogenicity, pigment biosynthesis, and several other processes (9). Quinolones and autoinducer 2 (AI-2) (51) also work as quorum-sensing molecules in some Gram-negative and Gram-positive bacteria. Similarly, in Gram-positive actinomycetes, antibiotic biosynthesis and differentiation are controlled by γ-butyrolactones or other autoinducers, e.g., PI factor (32), whereas Bacillus and Gram-positive cocci (1) have quorum-sensing mechanisms mediated by small peptides (22). Peptides also act as morphogens in Streptomyces griseus (45) and Streptomyces coelicolor (27). In plants, jasmonic acid is a well-known autoinducer (16). Autoinducers are very likely to occur in fungi, although so far little effort has been made to identify them. P. chrysogenum and A. chrysogenum produce extracellular peptides that might be autoinducers, as occurs in Bacillus and Gram-positive cocci.
Improvement of penicillin production in P. chrysogenum has been largely achieved by blind mutations that have resulted in amplification of chromosomal regions containing the pen gene cluster (10, 11, 48). Some external aromatic acids, such as phenylacetic acid or phenoxyacetic acid, increase production of specific penicillins (phenylacetyl- or phenoxyacetylpenicillins) by a precursor effect (21), but these aromatic acids are not autoinducers, since they are external precursors not synthesized by P. chrysogenum. Unlike external elicitors (2, 3, 13), autoinducers are synthesized by the producer organisms.
A molecule, conidiogenone, synthesized by Penicillium cyclopium is known to control differentiation in this fungus, and it may also affect the biosynthesis of secondary metabolites (34–36). Another fungal autoinducer is the butyrolactone I produced by Aspergillus terreus. Butyrolactone I [(α-oxo-β-(p-hydroxyphenyl)-γ-(p-hydroxy-m-3,3-dimethylallyl-benzyl)-γ- methoxycarbonyl-γ-butyrolactone)] contains the same nucleus as the Streptomyces γ-butyrolactones, although it belongs to a different biosynthetic class. Exogenous addition of butyrolactone I to the producer strain causes an increase in the production of several metabolites, particularly the anticholesterolemic lovastatin. In addition, butyrolactone I reduced colony growth and resulted in submerged spore formation (39).
The inducer 1,3-diaminopropane (1,3-DAP) described in this article is a rare low-molecular-weight diamine. There are three polyamines in eukaryotic cells, putrescine (1,4-diaminobutane), spermidine (1,8-diamino-4-azaoctano), and spermine (1,12-diamino-4,9-diazooctano). These are all aliphatic amines larger than 1,3-DAP. The latter compound is not frequently found in a free form. However, 1,3-DAP forms part of spermidine and is synthesized as an aminopropyl precursor for the biosynthesis of this diamine (see below).
The amino groups of 1,3-DAP and other diamines are protonated at physiological pH, and therefore these molecules are charged positively and interact with nucleic acids in the cells (37, 47). Polyamines are known to modulate binding of certain proteins (e.g., estrogen receptors) to Z-DNA-forming regions (44). Polyamines alter the specific DNA-protein interactions, particularly that of heat shock proteins and activator proteins induced by heat shock. Therefore, it is likely that 1,3-DAP modulates the expression of penicillin biosynthesis genes by interacting with the DNA and other activator proteins in the promoters of the pcbAB, pcbC, and penDE genes. The polyamines also participate in the stabilization of nucleosomes, and expression of penicillin and other secondary metabolite genes is influenced by heterochromatin organization (19, 40). It cannot be ruled out that the enhanced biomass formation in the presence of 1,3-DAP may also be related to an increase in fungal metabolic activity at early time points that may change the intracellular distribution of metabolites. This may facilitate growth and penicillin production.
The 1,3-DAP molecule derives from an aminopropyl group that originates by cleavage of S-adenosylmethionine (SAM), an activated form of methionine. In different eukaryotic organisms, SAM is first decarboxylated by a SAM decarboxylase, and the decarboxy-SAM is cleaved, generating an aminopropyl residue and methylthioadenosine. We have found the SAM decarboxylase gene (Pc21g01250) in the genome of P. chrysogenum (49). The second amino group of 1,3-DAP derives from one of the amino groups of putrescine, which in turn originates from ornithine by the action of the ornithine decarboxylase (ODC). Indeed, P. chrysogenum contains two putative genes for ornithine decarboxylase (Pc13g07510 and Pc21g18390). The condensation of the aminopropyl group with putrescine forms spermidine, in which the aminopropyl residue is bound to one of the amino groups of putrescine. The condensing enzyme named spermidine synthase has also been found by our group in P. chrysogenum (Pc13g12400).
It is very interesting that spermidine, unlike putrescine and spermine, also had a positive effect on penicillin production. This fact can be explained through the so-called retroconversion pathway described many years ago for Neurospora crassa (43). This pathway is involved in the cleavage of spermidine releasing 1,3-DAP and the five-carbon aminoaldehyde of putrescine (52). This cleavage is performed by a polyamine oxidase. We have found three polyamine oxidase genes in P. chrysogenum (Pc18g02250, Pc12g08750, and Pc22g02950). The 1,3-DAP released by the polyamine oxidase is secreted by a still-unknown mechanism, and as reported in this work, extracellular 1,3-DAP induces the expression of three genes involved in penicillin biosynthesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the European Union (Eurofungbase Consortium). Silvia M. Albillos was supported by a Torres Quevedo Contract from the Ministry of Science and Innovation of Spain (PTQ06-2-0113).
We thank M. J. López Nieto (Genhelix, Leon, Spain) for his support with the initial assays. We acknowledge the excellent technical assistance of B. Martín, J. Merino, A. Casenave, and A. Mulero.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Alloing G., Martin B., Granadel C., Claverys J. P. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75–83 [DOI] [PubMed] [Google Scholar]
- 2. Ariyo B. T., Bucke C., Keshaverz T. 1997. Alginate oligosaccharides as enhancers of penicillin production in cultures of Penicillium chrysogenum. Biotechnol. Bioeng. 53:17–20 [DOI] [PubMed] [Google Scholar]
- 3. Ariyo B. T., Tamerler C., Bucke C., Keshavarz T. 1998. Enhanced penicillin production by oligosaccharides from batch cultures of Penicillium chrysogenum in stirred-tank reactors. FEMS Microbiol. Lett. 166:165–170 [DOI] [PubMed] [Google Scholar]
- 4. Brakhage A. A. 1998. Molecular regulation of beta-lactam biosynthesis in filamentous fungi. Microbiol. Mol. Biol. Rev. 62:547–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casqueiro J., Bañuelos O., Gutiérrez S., Hijarrubia M. J., Martín J. F. 1999. Intrachromosomal recombination between direct repeats in Penicillium chrysogenum: gene conversion and deletion events. Mol. Gen. Genet. 261:994–1000 [DOI] [PubMed] [Google Scholar]
- 6. Chen X., et al. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549 [DOI] [PubMed] [Google Scholar]
- 7. Díez B., et al. 1990. The cluster of penicillin biosynthetic genes. J. Biol. Chem. 265:16358–16365 [PubMed] [Google Scholar]
- 8. Enríquez L. A., Pisano M. A. 1979. Isolation and nature of intracellular alpha-aminoadipic acid-containing peptides from Paecilomyces persicinus P-10. Antimicrob. Agents Chemother. 8:638–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Federle M. J., Bassler B. L. 2003. Interspecies communication in bacteria. J. Clin. Invest. 112:1291–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fierro F., et al. 1995. The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences. Proc. Natl. Acad. Sci. U. S. A. 92:6200–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fierro F., et al. 2006. Transcriptional and bioinformatic analysis of the 56.8 kb DNA region amplified in tandem repeats containing the penicillin gene cluster in Penicillium chrysogenum. Fungal Genet. Biol. 43:618–629 [DOI] [PubMed] [Google Scholar]
- 12. Galagan J. E., et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 13. Gang L., et al. 2001. Elicitation of penicillin biosynthesis by alginate in Penicillium chrysogenum exerted on pcbAB, pcbC, and penDE genes at transcriptional level. J. Ind. Microbiol. Biotechnol. 11:812–818 [Google Scholar]
- 14. García-Estrada C., Vaca I., Lamas-Maceiras M., Martín J. F. 2007. In vivo transport of the intermediates of the penicillin biosynthetic pathway in tailored strains of Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 76:169–182 [DOI] [PubMed] [Google Scholar]
- 15. Horinouchi S., Beppu T. 1992. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 46:377–398 [DOI] [PubMed] [Google Scholar]
- 16. Ishihara A., Ogura Y., Tebayashi S., Iwamura H. 2002. Jasmotane-induced changes in flavonoid metabolism in barley (Hordeum vulgare) leaves. Biosci. Biotechnol. Biochem. 66:2176–2182 [DOI] [PubMed] [Google Scholar]
- 17. Kleerebezem M., Quadri L. E. N., Kupers O. P., de Vos W. M. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895–904 [DOI] [PubMed] [Google Scholar]
- 18. Kosalková K., et al. 2000. A novel heptameric sequence (TTAGTAA) is the binding site for a protein required for high level expression of pcbAB, the first gene of the penicillin biosynthesis in Penicillium chrysogenum. J. Biol. Chem. 275:2423–2430 [DOI] [PubMed] [Google Scholar]
- 19. Kosalková K., et al. 2009. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 91:214–225 [DOI] [PubMed] [Google Scholar]
- 20. Laich F., Fierro F., Cardoza R. E., Martín J. F. 1999. Organization of the gene cluster for biosynthesis of penicillin in Penicillium nalgiovense and antibiotic production in cured dry sausages. Appl. Environ. Microbiol. 65:1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lamas-Maceiras M., Vaca I., Rodríguez E., Casqueiro J., Martín J. F. 2006. Amplification and disruption of the phenylacetyl-CoA ligase gene of Penicillium chrysogenum encoding an aryl-capping enzyme that supplies phenylacetic acid to the isopenicillin N acyltransferase. Biochem. J. 395:147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lazazzera B. E., Kurtser I. G., McQuade R. S., Grossman A. D. 1999. An autoregulatory circuit affecting peptide signalling in Bacillus subtilis. J. Bacteriol. 181:5193–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loder P. B., Abraham E. P. 1971. Isolation and nature of intracellular peptides from a cephalosporin C-producing Cephalosporium sp. Biochem. J. 123:471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. López-Nieto M. J., Ramos F. R., Luengo J. M., Martín J. F. 1985. Characterization of the biosynthesis in vivo of α-aminoadipyl-cysteinyl-valine in Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 22:343–351 [Google Scholar]
- 25. Machida M., et al. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161 [DOI] [PubMed] [Google Scholar]
- 26. Martín J. F., Ullán R. V., García-Estrada C. 2010. Regulation and compartmentalization of β-lactam biosynthesis. Microb. Biotechnol. 3:285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nodwell J. R., McGovern K., Losick R. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881–893 [DOI] [PubMed] [Google Scholar]
- 28. Pel H. J., et al. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221–231 [DOI] [PubMed] [Google Scholar]
- 29. Queener S. W., Ingolia T. D., Skatrud P. L., Chapman J. L., Kaster K. R. 1985. A system for genetic transformation of Cephalosporium acremonium, p. 468–472 In Leive L. (ed.), Microbiology. American Society for Microbiology, Washington, DC [Google Scholar]
- 30. Ramos F. R., López-Nieto M. J., Martín J. F. 1986. Coordinate increase of isopenicillin N synthetase, isopenicillin N epimerase and deacetoxycephalosporin C synthetase in a high cephalosporin-producing mutant of Acremonium chrysogenum and simultaneous loss of the three enzymes in a non-producing mutant. FEMS Microbiol. Lett. 35:123–127 [Google Scholar]
- 31. Ramsden M., McQuade B. A., Saunders K., Turner M. K., Harford S. 1989. Characterization of a loss-of-function mutation in the isopenicillin N synthetase gene of Acremonium chrysogenum. Gene 85:267–273 [DOI] [PubMed] [Google Scholar]
- 32. Recio E., Colinas A., Rumbero A., Aparicio J. F., Martín J. F. 2004. PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J. Biol. Chem. 279:41586–41593 [DOI] [PubMed] [Google Scholar]
- 33. Robson N. D., Cox A. R., McGowan S. J., Bycroft B. W., Salmond G. P. 1997. Bacterial N-acyl-homoserine-lactone-dependent signaling and its potential biotechnological applications. Trends Biotechnol. 15:458–464 [DOI] [PubMed] [Google Scholar]
- 34. Roncal T., Cordobés S., Sterner O., Ugalde U. 2002. Conidiation in Penicillium cyclopium is induced by conidiogenone, an endogenous diterpene. Eukaryot. Cell 1:823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roncal T., Cordobés S., Ugalde U., He Y., Sterner O. 2002. Novel diterpenes with potent conidiation activity. Tetrahedron Lett. 43:6799–6802 [Google Scholar]
- 36. Roncal T., Ugalde U. 2003. Conidiation induction in Penicillium. Res. Microbiol. 154:539–546 [DOI] [PubMed] [Google Scholar]
- 37. Ruiz Herrera J. 1994. Polyamines. DNA methylation and fungal differentiation. Crit. Rev. Microbiol. 20:143–150 [DOI] [PubMed] [Google Scholar]
- 38. Scherlach K., Herweck C. 2009. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 7:1753–1760 [DOI] [PubMed] [Google Scholar]
- 39. Schimmel T. G., Coffman A. D., Parsons S. H. 1998. Effect of butyrolactone I on the producing fungus, Aspergillus terreus. Appl. Environ. Microbiol. 64:3707–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shwab E. K., Keller N. P. 2008. Regulation of secondary metabolite production in filamentous ascomycetes. Mycol. Res. 112:225–230 [DOI] [PubMed] [Google Scholar]
- 41. Shen Y. Q., Wolfe S., Demain A. L. 1986. Levels of isopenicillin N synthetase and deacetoxycephalosporin C synthetase in Cephalosporium acremonium producing high and low levels of cephalosporin C. Biotechnology (NY) 4:61–64 [Google Scholar]
- 42. Shirafuji H., Fujisawa Y., Kida M., Kanzaki T., Yoneda M. 1979. Accumulation of tripeptide derivatives by mutants of Cephalosporium acremonium. Agric. Biol. Chem. 43:155–160 [Google Scholar]
- 43. Tabor C. W., Tabor H. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomas T., Thomas T. J. 1993. Structural specificity of polyamines in modulating the binding of estrogen receptor to potential Z-DNA forming sequences. J. Recept. Res. 13:1115–1133 [DOI] [PubMed] [Google Scholar]
- 45. Ueda K., et al. 2002. AmfS, an extracellular peptidic morphogen in Streptomyces griseus. J. Bacteriol. 184:1488–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ullán R. V., Campoy S., Casqueiro J., Fernández F. J., Martín J. F. 2007. Deacetylcephalosporin C production in Penicillium chrysogenum by expression of the isopenicillin N epimerization, ring expansion and acetylation genes. Chem. Biol. 14:329–339 [DOI] [PubMed] [Google Scholar]
- 47. van Dam L., Korolev N., Nordenskiöld L. 2002. Polyamine-nucleic acid interactions and the effects on structure in oriented DNA fibers. Nucleic Acids Res. 30:419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van den Berg M. A., Westerlaken I., Leeflang C., Kerkman R., Bovenberg R. A. 2007. Functional characterization of the penicillin biosynthetic gene cluster of Penicillium chrysogenum Wisconsin54-1255. Fungal Genet. Biol. 44:830–844 [DOI] [PubMed] [Google Scholar]
- 49. van den Berg M. A., et al. 2008. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 26:1161–1168 [DOI] [PubMed] [Google Scholar]
- 50. Wortman J. R., et al. 2009. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet. Biol. 46(Suppl. 1):S2–S13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xavier K. B., Bassler B. L. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamada H., Kimiyasu I., Yoshiki T. 1980. Oxidation of polyamines by fungal enzymes. Agric. Biol. Chem. 44:2469–2476 [Google Scholar]
- 53. Yamada Y., Nihira T. 1999. Microbial hormones and microbial chemical ecology, p. 377–413 In Barton S. D., Nakanishi K., Meth-Cohn O. (ed.), Comprehensive natural products chemistry, vol. 8 Elsevier, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.