Abstract
Since 2004, extended-spectrum cephalosporin (ESC)-resistant Salmonella enterica serovar Typhimurium (S. Typhimurium) isolates have been detected from cattle in the northern major island of Japan, Hokkaido. Resistance to ESCs was found to be mediated by CMY-2 type β-lactamase among 22 epidemiologically unrelated isolates showing indistinguishable pulsed-field gel electrophoresis patterns. Southern blot analysis using I-CeuI-digested genomic DNA demonstrated that the CMY-2 β-lactamase gene (blaCMY-2) was integrated in a 2.5-Mb chromosomal fragment. Genetic analysis of S. Typhimurium isolate L-3553 indicated that blaCMY-2 was located on a unique 125-kb genomic island, GI-VII-6, which consists of 140 open reading frames. Pairwise alignment of GI-VII-6 and Escherichia coli plasmid pAR060302 (size, 167 kb) revealed that a large proportion of GI-VII-6 (99%) shows a high sequence similarity (>99%) with pAR060302. GI-VII-6 contains 11 antimicrobial resistance genes including sul1, qacEΔ1, aadA2, and dfrA12 in the aadA2 region; sugE1 and blaCMY-2 in the blaCMY-2 region; and sul2, strA, strB, tet(A), and floR in the floR region. Two directly repeated IS26 copies were present at both ends of GI-VII-6. Junction regions of GI-VII-6 were flanked by an 8-bp direct repeat, indicating that GI-VII-6 was acquired by transposition involving IS26 transposase. PCR scanning revealed that the overall structure of GI-VII-6 was almost identical in the 22 isolates. Phylogenetic analysis suggested that S. Typhimurium isolates harboring GI-VII-6 belong to a different genomic lineage than other whole-genome-sequenced S. Typhimurium strains. These data indicate that a particular clone of S. Typhimurium harboring GI-VII-6 has spread among the cattle population in Hokkaido, Japan.
INTRODUCTION
Nontyphoidal salmonellae are a major cause of bacterial food-borne diseases worldwide (20). Typically, nontyphoidal salmonellae cause self-limiting gastroenteritis that does not require antimicrobial therapy (4). However, antimicrobial treatment is lifesaving for invasive salmonellosis, particularly in neonates of <1 year of age. Preventative antimicrobial treatment is also generally given to patients of >50 years of age who may have underlying atherosclerotic lesions that could be seeded by bacteremia (14). The options of first-line therapy for Salmonella infection include ampicillin, sulfamethoxazole-trimethoprim, fluoroquinolones, and extended-spectrum cephalosporins (ESCs). Among them, ESCs are preferred drugs, particularly in children requiring effective chemotherapy, because fluoroquinolones are not approved by the U.S. Food and Drug Administration for use in children in addition to the fact that resistance to ampicillin and sulfamethoxazole-trimethoprim is common in Salmonella enterica (S. enterica) isolates (14). Therefore, the emergence of ESC-resistant S. enterica is a global health problem (3).
Resistance to ESCs among S. enterica serovars has been attributed to the acquisition of plasmid-mediated β-lactamases, which are mostly Ambler class A extended-spectrum β-lactamases (ESBLs) and seldom Ambler class C cephamycinases (AmpC β-lactamases) (6). Of these, CMY-2, an AmpC β-lactamase encoded by the blaCMY-2 gene, is the most frequently detected enzyme (3). The blaCMY-2 gene has been horizontally transmitted through plasmids in various S. enterica serovars (12, 33, 34). Plasmid-mediated CMY-2 β-lactamase-producing S. enterica serovar Typhimurium (S. Typhimurium) isolates derived from cattle were reported for the first time in Japan in 2011 (29).
S. Typhimurium is a common cause of salmonellosis in humans and animals. Bovine salmonellosis is a dominant cause of morbidity and mortality in cattle and poses a threat to the beef and dairy industries (35). During the 1990s, S. Typhimurium definitive phage type 104 (S. Typhimurium DT104), having a core pattern of resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracyclines, emerged and was transmitted to many countries. S. Typhimurium DT104 was first detected in the United Kingdom and spread nearly simultaneously in North America, Europe, and Asia (13, 26, 30, 32). Surveillance data in Japan show that the incidence of S. Typhimurium DT104 bovine cases was stable until 1991 but increased over the next 3 years; then, the incidence of cases stabilized and even declined after 1995 (31).
The replacement of pentaresistant S. Typhimurium DT104 by a novel group of genetically related multidrug-resistant (MDR) S. Typhimurium isolates showing pulsed-field gel electrophoresis (PFGE) pattern cluster VII was noted in the major northern island of Japan, Hokkaido, during the 2000s (31). Between 2004 and 2006, a distinct clone of S. Typhimurium showing the specific PFGE pattern VII-6 (ST-VII-6, where ST is S. Typhimurium) and comprising cefazolin-resistant isolates was observed circulating among cattle herds in Hokkaido, Japan. Notably, a recent review of antimicrobial susceptibility interpretation criteria by the Clinical and Laboratory Standards Institute (CLSI) (10) reclassified this clonal group as ESC-resistant S. Typhimurium exhibiting resistance against third-generation cephalosporins. Moreover, preliminary phenotypic test results indicated that these isolates were potential cephamycinase producers, whereas resistance transfer experiments revealed that Escherichia coli transformants were susceptible to ESCs. Thus, these preliminary findings indicated the probability that the resistance determinant is located on the chromosome. Conversely, no reports so far have characterized chromosomally encoded blaCMY-2-producing S. Typhimurium isolates.
In the present study, we examined ST-VII-6 isolates derived from affected cattle for the following purpose: (i) to determine the genetic basis for antimicrobial resistance, (ii) to elucidate the location of the blaCMY-2 gene, and (iii) to manifest the gene arrangement of the blaCMY-2-flanking regions as a genomic island (GI).
MATERIALS AND METHODS
Bacterial isolation, identification, and typing.
The ST-VII-6 isolates examined in this study are listed in Table 1. These isolates were derived from affected cattle originating from different farms in Hokkaido, Japan, between 2004 and 2006. The isolation was performed by the staff of local Animal Hygiene Service Centers for the diagnostic purposes. Isolate L-3607 is the same as isolate KT262 described elsewhere (31). Salmonella spp. were identified based on colony morphology on selective medium and biochemical testing, as previously described (11). Serovar identification was performed by microtiter and slide agglutination methods according to the latest version of the Kaufmann and White scheme (24) using antiserum (Denka Seiken, Tokyo, Japan). Phage typing of the prototype S. Typhimurium isolate L-3553 was performed according to the methods and schemes previously described by Anderson et al. (2). PFGE typing using XbaI endonuclease (Takara Bio, Shiga, Japan) was performed according to the PulseNet protocol (25) and interpreted as previously described (31). These isolates were maintained at −80°C in Trypto-Soya broth (Nissui Pharmaceutical, Tokyo, Japan) containing 25% (vol/vol) glycerol.
Table 1.
S. Typhimurium isolates showing PFGE pattern cluster VII-6 used in this study
| Isolatea | Year of isolation | Antimicrobial resistance profileb |
|---|---|---|
| L-3553–L-3554 | 2004 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
| L-3556 | 2004 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
| L-3593 | 2004 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
| L-3594 | 2005 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
| L-3595 | 2005 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, SUL, SXT |
| L-3597 | 2005 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
| L-3599–L-3600 | 2005 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
| L-3605–L-3608 | 2005 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
| L-3770–L-3775 | 2005 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
| L-3777–L-3779 | 2006 | AMP, CFZ, FOX, CAZ, CTX, KAN, STR, TET, CHL, SUL, SXT |
All isolates carried the 130-kb plasmid.
AMP, ampicillin; CFZ, cefazolin; FOX, cefoxitin; CAZ, ceftazidime; CTX, cefotaxime; KAN, kanamycin; STR, streptomycin; TET, tetracycline; CHL, chloramphenicol; SUL, sulfonamide; SXT, sulfamethoxazole-trimethoprim.
Antimicrobial susceptibility testing.
The Kirby-Bauer disc diffusion test was performed using Mueller-Hinton agar plates (Becton, Dickinson, and Co., Sparks, MD) according to CLSI standards (formerly National Committee for Clinical and Laboratory Standards) (23) using the following antimicrobials: ampicillin (10 μg), cefazolin (30 μg), cefoxitin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), kanamycin (30 μg), streptomycin (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), fosfomycin (50 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), sulfamethoxazole (250 μg), nalidixic acid (30 μg), ofloxacin (5 μg), norfloxacin (10 μg), and ciprofloxacin (5 μg) (Becton, Dickinson, and Co.). Susceptibility testing results in this study were interpreted according to the new criteria established by CLSI (10).
Cephalosporinase production by S. Typhimurium isolates was detected by the P/Case test (Nissui Pharmaceutical) according to the manufacturer's instructions.
PCR.
A single colony of each bacterial isolate was suspended in 50 μl of 25 mM NaOH and boiled for 5 min. After addition of 4 μl of 1 M Tris-HCl (pH 8.0), the suspension was centrifuged, and the supernatant was used as template DNA. Oligonucleotides were purchased from Hokkaido System Science (Sapporo, Japan). Amplification was performed using an iCycler apparatus (Bio-Rad Laboratories, Hercules, CA). Takara Ex Taq and Takara LA Taq (Takara Bio) were used as DNA polymerases to amplify fragments in sizes of <7 kb and >7 kb, respectively, according to the manufacturer's instructions.
Plasmid isolation and transformation.
Plasmid DNA was isolated by the method described by Kado and Liu (16) followed by phenol-chloroform extraction. A Bac-Tracker Supercoiled DNA ladder (Epicentre Biotechnologies, Madison, WI) was used as a size marker. Transformation of E. coli DH5α was performed by electroporation using a MicroPulser (Bio-Rad Laboratories) according to the manufacturer's instructions. The transformants were selected on Luria-Bertani agar plates (Becton, Dickinson, and Co.) containing ampicillin (25 μg/ml) or cefazolin (25 μg/ml).
Localization of resistance genes by Southern blot analyses.
To confirm the chromosomal location of blaCMY-2 and associated resistance genes, plugs for I-CeuI restriction were prepared from the total DNA of three selected representative isolates (L-3553, L-3607, and L-3777) and from S. Typhimurium strain LT2 (LT2) according to the PulseNet protocol (25), with a few modifications. In brief, the cell culture for plug preparation had an optical density at 600 nm of 2, the final concentration of proteinase K in the plugs was 1 mg/ml, and one additional wash (each) in distilled water and in Tris-EDTA (TE) buffer was done. The plugs were digested with 40 U/ml of I-CeuI enzyme (New England BioLabs, Ipswich, MA) at 37°C for 4 h. The digested fragments were separated by PFGE (6.0 V/cm for 8 h and 12 h with a pulsing time linearly ramped from 5 to 20 s and 20 to 70 s, respectively) on a 1% agarose gel. The sizes of restriction fragments were determined relative to known I-CeuI fragment sizes of LT2 (18). The plugs for XbaI restriction (Takara Bio) were prepared, digested, and separated according to the PulseNet protocol (25). Separated I-CeuI and XbaI fingerprints were transferred to Hybond-N+ nylon membranes (Amersham Biosciences, Buckingham, United Kingdom) and hybridized with PCR-generated digoxigenin-labeled probes (Roche Diagnostics Co., Indianapolis, IN) (see Table S1 in the supplemental material) using PerfectHyb hybridization solution (Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer's instructions.
Southern blot analysis was performed using S1 nuclease-digested genomic DNA to elucidate the plasmid contents of the isolates. In brief, total DNA in the plugs was treated with 2 U/ml of S1 nuclease (Takara Bio) and incubated at 37°C for 45 min. The plasmid bands were separated by PFGE (6.0 V for 20 h with a pulsing time linearly ramped from 5 s to 50 s) and hybridized using digoxigenin-labeled gene probes as described above.
Short-read DNA sequencing and de novo assembly.
An approximately 500-bp DNA library of S. Typhimurium isolate L-3553 was prepared using a genomic DNA Sample Prep kit (Illumina, San Diego, CA), and DNA clusters were generated on a slide using a Cluster Generation kit (version 4) on an Illumina cluster station, according to the manufacturer's instructions. In brief, to obtain approximately 1.0 × 107 clusters for a single lane, the following series of reactions were performed using standard Illumina protocols: template hybridization, isothermal amplification, linearization, blocking, denaturation, and hybridization of the sequencing primer. All sequencing runs for 80-mers were performed using an Illumina Genome Analyzer II (GA II) using a TruSeq SBS kit (version 5). Fluorescent images were analyzed using the Illumina base-calling pipeline RTA1.8/SCS2.8 to obtain FASTQ-formatted sequence data. Before de novo assembly, obtained 80-mer reads were assembled using ABySS-pe, version 1.25 (27), with setting parameters (k-mer, 40; minimum number of pairs needed to consider joining two configs, 70; minimum coverage for assembly, 50).
DNA sequencing of the GI.
Our preliminary unpublished data suggest that blaCMY-2 was integrated in the chromosome with elements of plasmid DNA, similar to E. coli plasmid pAR060302 (size, 167 kb) (7). Therefore, we attempted PCR amplification of approximately 11-kb fragments from the genomic DNA of L-3553 using the sequence data of pAR060302 as a reference, and then we determined the DNA sequence of each fragment by Sanger DNA sequencing using a BigDye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems, Foster City, CA). The predicted amplified fragments contain overlaps to assemble with flanking fragments. To identify the integration site of the GI on the chromosomal DNA, a BLASTN homology search (1) was performed using the possible junction sequence of the GI as a query against all obtained de novo assemblies. Predicted gaps between contigs were amplified using specific PCR primer pairs, followed by Sanger DNA sequencing. To determine if misassembled sequences and incorrect gap closing persisted after the sequencing analysis of the GI, 40-mer short reads were aligned to the tentative complete sequence of the GI with Maq software (version 0.7.1) using the “easyrun” Perl command (17). Rapid alignment to validate possible errors was performed using a MapView graphical alignment viewer (5).
Pairwise alignment of the GI with relevant plasmid sequences.
Pairwise alignment of the GI element was performed by BLASTN homology search with the GI element of relevant plasmids, followed by visualization of the aligned images using the ACT program (8).
DNA sequencing of antimicrobial resistance regions in a 130-kb plasmid of L-3553.
Mapping results of short DNA reads obtained from isolate L-3553 using the sequence data of S. Typhimurium plasmid pU302L (9) as a reference suggested that the isolate contained antimicrobial resistance genes homologous to those of pU302L. Therefore, predicted resistance regions were amplified using specific PCR primer pairs, listed in Table S2 in the supplemental material, followed by Sanger DNA sequencing using a BigDye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems).
Annotation.
Open reading frames (ORFs) were predicted and assigned using a BLASTN homology search for GI sequence data and antimicrobial resistance regions in a 130-kb plasmid of isolate L-3553.
Analysis of antimicrobial resistance regions in other S. Typhimurium isolates.
To determine the distribution of the GI among S. Typhimurium isolates, we established a system, here referred to as PCR scanning, to amplify an entire sequence of the GI. Table S3 in the supplemental material shows a list of primers used for PCR amplification of 19 segments of the GI whereby each segment contains overlaps in both ends with flanking segments. To elucidate the structure of each resistance region, we established another system, here referred to as PCR mapping. Table S4 in the supplemental material shows a list of primers employed to amplify DNA fragments containing the 5′ and/or 3′ ends of each resistance gene located in the chromosome or 130-kb plasmid (see Fig. S1 in the supplemental material).
Extraction of SNVs from core genome sequences among S. Typhimurium strains.
Single nucleotide variations (SNVs) of isolate L-3553 were identified and compared to the reference chromosomal sequence of LT2 (GenBank accession number AE006468) using Maq software (version 0.7.1) (17) with the easyrun command as the default parameter. Strain-specific SNVs were extracted from the cns.final.snp files. Whole SNVs were extracted from the available genomic sequences of other S. Typhimurium strains using Maq software (version 0.7.1), and the “Maq simulate” command was used with the following modifications of the default parameters: number of pairs of reads, 10,000,000; mutation rate, 0; and fraction of 1-bp indels, 0. These parameters indicate that 20 million 35-mer hypothetical reads were generated without mutations or indels from the genomic sequences used for SNV identification. SNVs located in prophages (Fels-1, Fels-2, Gifsy-1, and Gifsy-2) (21) were excluded from further phylogenetic analysis.
Phylogenetic analysis.
All SNVs were concatenated to generate a pseudo-sequence for phylogenetic analysis. A DNA maximum-likelihood program (RAxML, version 7.25) (28) was used for phylogenetic analysis by bootstrapping 1,000 times. FigTree software, version 1.2.3, was used to display the generated tree.
MLST analysis.
The sequence type (ST) of isolate L-3553 was determined at the multilocus sequence typing (MLST) website for S. enterica (http://mlst.ucc.ie/mlst/mlst/dbs/Senterica/). Nucleic acid regions for seven loci used for MLST were extracted from the de novo assemblies and deposited to the website.
Nucleotide sequence accession numbers.
The nucleotide sequences of S. Typhimurium isolate L-3553 were deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers DRA000367 for the short reads, AB571791 for the GI, and AB571792 to AB571795 for the resistance regions of the 130-kb plasmid, pST3553.
RESULTS
Phenotypic characteristics of ST-VII-6 isolates.
The antimicrobial resistance phenotypes of ST-VII-6 isolates are shown in Table 1. All 22 isolates exhibited resistance to β-lactams including ampicillin, cefazolin, cefoxitin, ceftazidime, and cefotaxime. In addition, all isolates showed resistance to kanamycin, streptomycin, tetracycline, chloramphenicol, sulfonamide, and sulfamethoxazole-trimethoprim, except for isolate L-3595, which was susceptible to chloramphenicol. However, all isolates were susceptible to fosfomycin, nalidixic acid, ofloxacin, norfloxacin, and ciprofloxacin. The E. coli strains transformed by the 130-kb plasmid of ST-VII-6 exhibited resistance to ampicillin, kanamycin, and tetracycline. The P/Case test detected cephalosporinase production by ST-VII-6 isolates. Isolate L-3553 was phage untypeable with the standard Colindale panel of phages.
Localization of antimicrobial resistance genes.
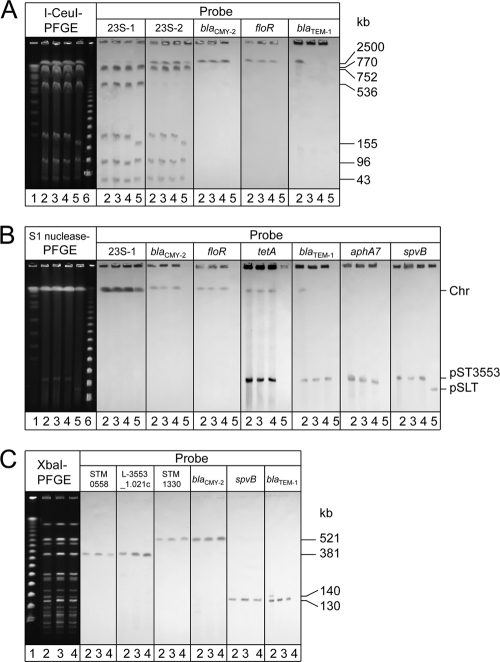
LT2 contains seven I-CeuI restriction sites located on the 23S rRNA (rrl) genes on the chromosome, and the orientation of two of the seven rrl genes is the opposite of that of the others. I-CeuI digestion of the LT2 chromosome generates seven fragments of sizes 2,500, 770, 752, 536, 155, 96, and 43 kb. To detect all seven fragments by Southern blot analysis, two probes, i.e., 23S-1 and 23S-2, that targeted rrl sequences upstream and downstream, respectively, from the I-CeuI restriction sites were employed. The 23S-1 probe hybridized six fragments of sizes 770, 752, 536, 155, 96, and 43 kb. On the other hand, the 23S-2 probe hybridized six fragments of sizes 2,500, 770, 752, 155, 96, and 43 kb, as expected. The blaCMY-2 gene probe hybridized with a 2.5-Mb fragment, as did the floR and tet(A) gene probes, confirming that the cephalosporinase, blaCMY-2, floR, and tet(A) genes were located on the chromosomes of the three examined isolates (Fig. 1A). In addition, the blaTEM-1 gene probe hybridized with the same chromosomal fragment only in isolate L-3553.
Fig. 1.
PFGE-Southern blot hybridization images demonstrating chromosomal and/or plasmid location of various genes. (A) PFGE separation of I-CeuI-digested genomic DNA from S. enterica serovar Typhimurium isolates followed by Southern blot hybridization with 23S-1 rRNA, 23S-2 rRNA, blaCMY-2, floR, and blaTEM-1 gene probes. Lane 1, CHEF DNA size standard; lane 2, isolate L-3553; lane 3, isolate L-3607; lane 4, isolate L-3777; lane 5, strain LT2; and lane 6, Lambda ladder marker. (B) PFGE separation of S1 nuclease-digested genomic DNA from S. Typhimurium isolates followed by Southern blot hybridization with 23S-1 rRNA, blaCMY-2, floR, tet(A), blaTEM-1, aphA7, and spvB gene probes. Lane 1, CHEF DNA size standard; lane 2, isolate L-3553; lane 3, isolate L-3607; lane 4, isolate L-3777; lane 5, strain LT2; and lane 6, Lambda ladder marker. Chr, chromosomal material. (C) XbaI-digested whole-genomic DNA from S. Typhimurium isolates, followed by Southern blot hybridization with STM0558, L-3553_1.021c, blaCMY-2, blaTEM-1, and spvB gene probes. Lane 1, Lambda ladder marker; lane 2, isolate L-3553; lane 3, isolate L-3607; and lane 4, isolate L-3777.
Southern blot analysis using genomic DNA after S1 nuclease digestion confirmed that both the blaCMY-2 and floR genes were absent in any of the plasmids including the 130-kb plasmid. In contrast, hybridization signals of the tet(A), blaTEM-1, aphA7, and spvB gene probes were present in the 130-kb plasmid of ST-VII-6 isolates (Fig. 1B). The probes for blaTEM-1 in isolate L-3553, in addition to blaCMY-2, floR, and tet(A) in representative isolates L-3553, L-3607, and L-3777, were hybridized with the chromosomal material; this further demonstrated the existence of chromosomal copies of these genes (Fig. 1B).
The spvB and blaTEM-1 gene probes were hybridized with a 130-kb fragment generated by XbaI digestion of genomic DNA from the representative isolates L-3553, L-3607, and L-3777, suggesting that the 130-kb fragment corresponds to pST3553 (Fig. 1C). A weak hybridization signal was also detected on a 140-kb fragment generated by XbaI digestion of genomic DNA from isolate L-3553 exhibiting existence of an extra chromosomal copy of blaTEM-1 in this isolate (Fig. 1C).
Structure and location of the GI in isolate L-3553.
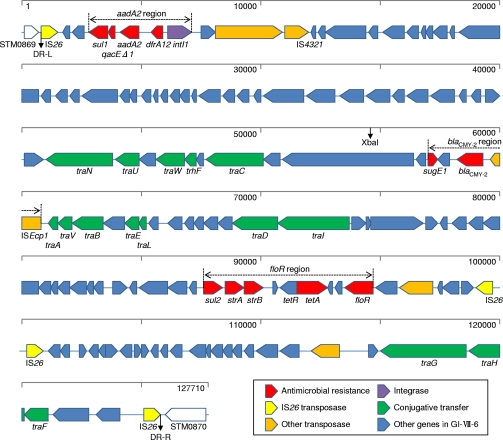
DNA sequencing analysis indicated that isolate L-3553 possesses a unique 125-kb genomic island, designated GI-VII-6, which consists of 140 ORFs. GI-VII-6 contains 10 antimicrobial resistance genes in addition to blaCMY-2 in three distinct resistance regions (Fig. 2; see also Table S5 in the supplemental material). The aadA2 region located near the left-terminal IS26 element comprises a class 1 integron, with the aadA2 and dfrA12 gene cassettes conferring resistance to streptomycin/spectinomycin and trimethoprim, respectively, followed by the 3′ conserved region of the class 1 integron composed of sul1 and qacEΔ1, conferring resistance to sulfonamides and quaternary ammonium compounds, respectively. The blaCMY-2 region comprises the gene array ISEcp1-blaCMY-2-blc1-sugE1. blc1 encodes the outer membrane lipoprotein, and sugE1 mediates the drug efflux channel/quaternary ammonium compound resistance protein. The floR region includes resistance genes sul2, strA, strB, tet(A), and floR and mediates resistance to sulfonamides, streptomycin, streptomycin, tetracyclines, and chloramphenicol/florfenicol, respectively.
Fig. 2.
Schematic view of full-length GI-VII-6 of S. Typhimurium isolate L-3553. The diagram shows the classification of each gene according to the following scheme: yellow, genes for IS26 transposase; red, resistance genes; orange, genes for other transposases; purple, gene for integrase; green, genes for conjugative transfer; blue, other genes in GI-VII-6. The three resistance regions shown are the aadA2 resistance region, the blaCMY-2 resistance region, and the floR resistance region. A distance scale in bp is indicated at the top of each map, and the coordinates are from the complete GI-VII-6 sequence (DDBJ accession number AB571791). An XbaI restriction site is shown.
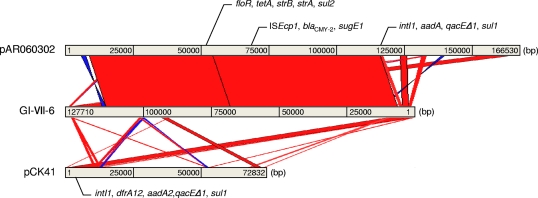
Pairwise alignment of GI-VII-6 and E. coli plasmid pAR060302 revealed a large proportion of GI-VII-6 (99%) sharing high sequence similarity (>99%) with pAR060302. The DNA sequence of an 8,781-bp region containing the aadA2 region (nucleotides [nt] 1201 to 9981) displayed 99% sequence similarity with the region of Edwardsiella tarda plasmid pCK41 at nt positions 18721 to 22692 (Fig. 3; see also Table S5 in the supplemental material). All type IV conjugative transfer genes of pAR060302 were found in GI-VII-6, whereas genes responsible for plasmid replication, such as ori and repA, were missing (Fig. 2; see also Table S5 in the supplemental material). Four of nine transposase genes located in GI-VII-6 were from IS26 (Fig. 2; see also Table S5), and directly repeated IS26 copies were located at both ends of GI-VII-6. In addition, GI-VII-6 was flanked by an 8-bp (CTCCACAA) direct repeat and was found to be inserted in the intergenic region between the STM0869 and STM0870 homologous genes of the corresponding LT2 (Fig. 2; see also Table S5 in the supplemental material).
Fig. 3.
Schematic representation of multidrug resistance determinants. Pairwise comparison of GI-VII-6 in S. Typhimurium isolate L-3553 with E. coli plasmid pAR060302 and E. tarda plasmid pCK41 by BLASTN homology search and visualized by the ACT program. The red and blue bars between the chromosomal DNA lines shown in gray represent individual nucleotide matches with forward or reverse direction, respectively. BLASTN match scores less than 200 are not shown.
The entire sequence of GI-VII-6 included a single XbaI restriction site (nt 54682 to 54687), which suggested that XbaI digestion of L-3553 genomic DNA would generate two different fragments containing a partial sequence of GI-VII-6 in one end (see Fig. S2 in the supplemental material). The predicted sizes of the fragments were 381 and 521 kb, based on the sequence data of LT2. ORF L-3553_1.021c in GI-VII-6 and the STM0558 homolog were believed to be present in the 381-kb fragment, whereas blaCMY-2 in GI-VII-6 and the STM1330 homolog were present in the 521-kb fragment. Positive results from the Southern blot hybridization of XbaI-digested DNA fragments using the probes prepared from the above target genes confirmed these hypotheses (Fig. 1C).
DNA sequences of antimicrobial resistance regions in pST3553.
Results of short-read mapping, using the pU302L sequence as a reference, indicated that isolate L-3553 consists of four antimicrobial resistance regions similar to those of pU302L. PCR and DNA sequencing revealed that these regions contained the resistance genes aphA7, blaTEM-1, blaTEM-1, and tet(A) (see Fig. S1A in the supplemental material). Moreover, these resistance regions were found to be outside the domain of GI-VII-6. Localization experiments by Southern blot analysis using S1 nuclease and XbaI digestions confirmed that these genes were actually located on pST3553 (Fig. 1B and C).
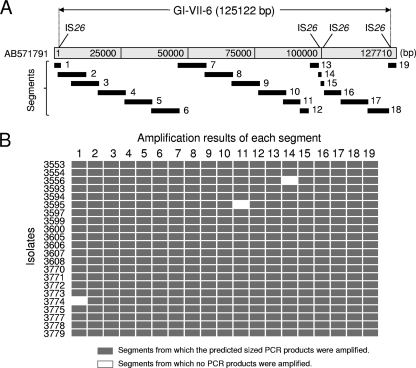
Prevalence of GI-VII-6 and antimicrobial resistance genes among ST-VII-6 isolates.
PCR scanning of GI-VII-6 in the ST-VII-6 isolates revealed that 19 of 22 isolates were PCR positive for all of the 19 segments, with expected product sizes (Fig. 4). Each of the remaining three isolates (L-3556, L-3595, and L-3774) exhibited one unamplified segment (Fig. 4B); i.e., the overall structure of GI-VII-6 was almost identical among the 22 isolates. Investigation of the structure of antimicrobial resistance regions in GI-VII-6 and a 130-kb plasmid by PCR mapping revealed that 21 of 22 ST-VII-6 isolates amplified all fragments with expected product sizes (see Fig. S1 in the supplemental material). On the other hand, fragments 14 and 15 were not amplified in the chloramphenicol-sensitive isolate L-3595. DNA sequence analysis in this isolate suggested that the floR gene was deleted by a single crossover between directly repeated 307-bp sequences at nt positions 92701 to 93007 and at 96985 to 97291 (data not shown).
Fig. 4.
Summary for PCR scanning of GI-VII-6 among 22 clonally related S. Typhimurium isolates. (A) Positions of segments designed to amplify 19 regions covering the full length of GI-VII-6. (B) Gray and white rectangles indicate segments from which the predicted size products were amplified or not amplified, respectively. The nucleotide sequence of S. Typhimurium L-3553 is available in the DDBJ (accession number AB571791).
Genomic information of isolate L-3553.
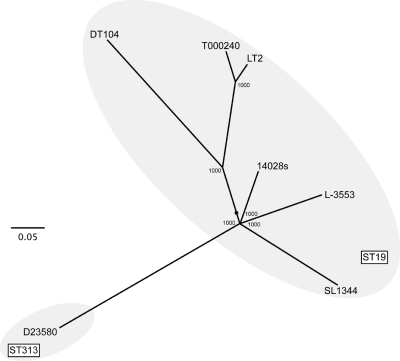
MLST analysis revealed that L-3553 belongs to ST19, which includes the S. Typhimurium strains LT2, T000240, DT104, 14028s, and SL1344. To further characterize the progeny of serovar S. Typhimurium strains, the 1,920 SNVs identified on the basis of a common shared genome sequence (see Table S6 in the supplemental material), excluding the prophages Fels-1, Fels-2, Gifsy-1, and Gifsy-2, were analyzed by the maximum-likelihood method. L-3553 contains 210 unique SNVs which are not shared by other strains. These data suggest that the genomic lineage of L-3553 is different from that of other ST19 strains (Fig. 5).
Fig. 5.
Phylogenetic tree based on core genome SNVs among whole-genome-sequenced S. Typhimurium strains by the maximum-likelihood method with 1,000-fold bootstrapping. The scale indicates that a branch length of 0.05 is five times as long as one that would show a 1% difference between the nucleotide sequences at the beginning and end of the branch. The number at each branch node represents the bootstrapping value. The ST19 and ST313 groups, as determined by MLST, are indicated by circles. The SNV information is summarized in Table S6 in the supplemental material.
DISCUSSION
In this study, we demonstrated that ST-VII-6 isolates carry a chromosomally encoded blaCMY-2 gene located on the GI involved in multidrug resistance, based on several observations. Southern blot analysis using selected isolates and various probes revealed that the blaCMY-2 gene was located on a 2.5-Mb fragment originating from chromosomal DNA (Fig. 1A), whereas S1 nuclease PFGE confirmed the absence of the gene on large plasmids including pST3553 (Fig. 1B). Moreover, DNA sequencing analysis of isolate L-3553 manifested that a single copy of blaCMY-2 was located on a unique 125-kb GI (Fig. 2; see Table S5 in the supplemental material), and the chromosomal location of blaCMY-2 was further confirmed by Southern blot analysis using XbaI-digested genomic DNA (Fig. 1C). PCR scanning results revealed that most ST-VII-6 isolates harbor an intact GI-VII-6 (Fig. 4). Conversely, we could not find any evidence indicating that isolate L-3553 possessed another copy of blaCMY-2 in the genome. Thus, to the best of our knowledge, this is the first report clearly demonstrating the chromosomal location of blaCMY-2 among ESC-resistant S. Typhimurium isolates originating from food-producing animals worldwide.
Evidence for the chromosomal and plasmid locations of blaCMY-2 in an S. Typhimurium isolate from a human clinical case was reported for the first time in 2008 in the United States (36). Zioga et al. conducted further studies to establish the role of the chromosomal copy of the blaCMY-2 gene in ESC resistance. However, they could not determine resistance levels attributed to the blaCMY-2 gene because a comparison of total cephalosporinase activities by UV spectrophotometry using cell extracts and cefalotin as a reporter substrate did not indicate any significant influence of the extra chromosomal copy (36). In our study, we could not detect CTX-M, TEM, and SHV ESBL gene types by PCR (data not shown) from the ST-VII-6 isolates, suggesting that resistance to ESCs was solely attributed to the chromosomally located blaCMY-2 gene. The type of β-lactamase gene contained in a 130-kb plasmid among ST-VII-6 isolates was identified as blaTEM-1, which confers resistance primarily to penicillins such as ampicillin.
A large proportion of GI-VII-6 shares a high degree of sequence similarity to the E. coli IncA/C plasmid pAR060302 originating from cattle in the United States (Fig. 3). IncA/C plasmids have been implicated as common carriers of the CMY-2 type cephalosporinase gene (7). This genetic relationship may indicate that plasmid DNA is a possible source of the GI-VII-6 backbone. Moreover, GI-VII-6 carries IS26 at both ends and is flanked by an 8-bp direct repeat because of target site duplication. The presence of a direct duplication of target sequence at each side of the MDR region indicates that the element was inserted between homologous genes of STM0869 and STM0870 by a transposition mechanism, as described previously for IS26 composite transposons (15, 19, 22). The IS26 is widespread among plasmids and could be implicated in the acquisition of resistance genes in different mechanisms (22). Thus, transposition and/or homologous recombination of IS26 associated with integrons and transposons is probably implicated in building the GI-VII-6 core structure manifested in this work. It could, therefore, represent the initial steps in the generation of more or less large IS26 composite transposons.
Recent movement of blaCMY-2 plasmids among E. coli and S. enterica hosts has been evidenced (7). In addition, Call et al. described the self-conjugation of IncA/C plasmid pAR060302 with relatively high efficiency (7). Therefore, the IncA/C blaCMY-2 plasmid backbone of GI-VII-6 was possibly horizontally transmitted by conjugation of the donor strain with an ST-VII-6 ancestor as a recipient using a type IV conjugative transfer system. This was followed by chromosomal integration of the blaCMY-2 plasmid backbone, probably through an IS26-mediated transposition mechanism. A related process was previously described involving members of the IS6 family (e.g., IS26), giving rise to replicon fusions (cointegrates) in which the donor and target replicons were separated by two directly repeated IS copies. Consequently, a resolution step was involved in separating donor and target replicons with subsequent transfer of the transposon to the target (19). Notably, previous studies revealed that Tn2680, a composite transposon, consisting of a kanamycin resistance gene surrounded by two IS26 copies, could mediate both transposition of the kanamycin resistance gene through cointegration of the entire plasmid carrying Tn2680 and subsequent resolution by homologous recombination between two IS26 elements (15). Interestingly, although the tra genes (encoding type IV conjugative transfer proteins) were highly conserved in GI-VII-6, the island did not appear to carry the ori and repA genes (Fig. 2; see also Table S5 in the supplemental material). The lack of these genes may be related to genetic rearrangements because of IS26 transposition events leading to the deletion of these genes during a resolution step required to separate donor and target DNA as previously described (19).
Analysis of the plasmid contents indicated a 130-kb plasmid, pST3553, showing a high degree of sequence similarity to pU302L from S. Typhimurium. Notably, pST3553 seems to be a virulence resistance plasmid because the mapping of short DNA reads revealed an 80,216-bp sequence of L-3553 matching the pSLT sequence (data not shown). In addition, S1 nuclease-PFGE and Southern blot hybridization using the spvB gene probe demonstrated positive hybridization signals with both pSLT and pST3553, indicating that pST3553 together with 130-kb plasmids carried by other isolates comprise the virulence plasmid (similar to pSLT) combined with another resistance determinant (Fig. 1B). Moreover, pST3553 carries two copies of the TEM-1 β-lactamase gene, and isolate L-3553 harbors an extra copy of the TEM-1 β-lactamase gene in the chromosome outside the domain of GI-VII-6. These multiple copies of the same resistance gene may increase antimicrobial resistance levels and provide more selective advantage.
Taken together, results of this study represent the first characterization of the unique genomic island GI-VII-6 involved in acquisition of the blaCMY-2 gene by S. Typhimurium clinical isolates originating from farm animals. Phylogenetic analyses revealed that ST-VII-6 isolates belong to a different genomic lineage from other whole-genome-sequenced S. Typhimurium strains. These results indicate that a particular clone of S. Typhimurium harboring GI-VII-6 has spread in the cattle population in Hokkaido, Japan. These findings also suggest that the blaCMY-2 gene may spread among members of Enterobacteriaceae family through chromosomal mobilization of the IncA/C plasmid backbone by IS26 cointegration and/or homologous recombination. The probability that this clone will spread is a public health concern because it may contribute to the amplification and persistence of blaCMY-2, limiting the effectiveness of cephalosporin therapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants-in-aid from the Ministry of Health, Labor, and Welfare of Japan (H21-Shokuhin-Ippan-013) and the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant number 22-00217).
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 27 June 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Anderson E. S., Ward L. R., Saxe M. J., de Sa J. D. 1977. Bacteriophage-typing designations of Salmonella Typhimurium. J. Hyg. (Lond.) 78:297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arlet G., et al. 2006. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945–1954 [DOI] [PubMed] [Google Scholar]
- 4. Aserkoff B., Bennett J. V. 1969. Effect of antibiotic therapy in acute salmonellosis on the fecal excretion of salmonellae. N. Engl. J. Med. 281:636–640 [DOI] [PubMed] [Google Scholar]
- 5. Bao H., et al. 2009. MapView: visualization of short reads alignment on a desktop computer. Bioinformatics 25:1554–1555 [DOI] [PubMed] [Google Scholar]
- 6. Bonnet R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Call D. R., et al. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carver T., et al. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen C. Y., Nace G. W., Solow B., Fratamico P. 2007. Complete nucleotide sequences of 84.5- and 3.2-kb plasmids in the multi-antibiotic resistant Salmonella enterica serovar Typhimurium U302 strain G8430. Plasmid 57:29–43 [DOI] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing, 20th information supplement M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Ewing W. H. 1986. Edwards and Ewing's identification of Enterobacteriaceae, 4th ed Elsevier Science Publishing Co., Inc., New York, NY [Google Scholar]
- 12. Giles W. P., et al. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob. Agents Chemother. 48:2845–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glynn M. K., et al. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333–1338 [DOI] [PubMed] [Google Scholar]
- 14. Hohmann E. L. 2001. Nontyphoidal salmonellosis. Clin. Infect. Dis. 32:263–269 [DOI] [PubMed] [Google Scholar]
- 15. Iida S. B., Meyer J., Arber W. 1984. Functional characterization of the prokaryotic mobile genetic element IS26. Mol. Gen. Genet. 198:84–89 [DOI] [PubMed] [Google Scholar]
- 16. Kado C. I., Liu S. T. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H., Ruan J., Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu S. L., Sanderson K. E. 1995. I-CeuI reveals conservation of the genome of independent strains of Salmonella Typhimurium. J. Bacteriol. 177:3355–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mahillon J., Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Majowicz S. E., et al. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50:882–889 [DOI] [PubMed] [Google Scholar]
- 21. McClelland M., et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 22. Miriagou V., Carattoli A., Tzelepi E., Villa L., Tzouvelekis L. S. 2005. IS26-associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrob. Agents Chemother. 49:3541–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. National Committee for Clinical Laboratory Standards 2001. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals, 2nd ed Approved standard M31-2A. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 24. Popoff M. Y. 2001. Antigenic formulas of the Salmonella serovars, 8th ed WHO Collaborating Centre for Reference and Research on Salmonella. Institut Pasteur, Paris, France [Google Scholar]
- 25. Ribot E. M., et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 26. Sameshima T., et al. 2000. Salmonella Typhimurium DT104 from livestock in Japan. Jpn. J. Infect. Dis. 53:15–16 [PubMed] [Google Scholar]
- 27. Simpson J. T., et al. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stamatakis A., Ludwig T., Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463 [DOI] [PubMed] [Google Scholar]
- 29. Sugawara M., et al. 2011. Molecular and phenotypic characteristics of CMY-2 beta-lactamase-producing Salmonella enterica serovar Typhimurium isolated from cattle in Japan. J. Vet. Med. Sci. 73:345–349 [DOI] [PubMed] [Google Scholar]
- 30. Tamada Y., et al. 2001. Molecular typing and epidemiological study of Salmonella enterica serotype Typhimurium isolates from cattle by fluorescent amplified-fragment length polymorphism fingerprinting and pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamamura Y., et al. 2011. Molecular epidemiology of Salmonella enterica serovar Typhimurium isolates from cattle in Hokkaido, Japan: evidence of clonal replacement and characterization of the disseminated clone. Appl. Environ. Microbiol. 77:1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Threlfall E. J., Frost J. A., Ward L. R., Rowe B. 1994. Epidemic in cattle and humans of Salmonella Typhimurium DT 104 with chromosomally integrated multiple drug resistance. Vet. Rec. 134:577. [DOI] [PubMed] [Google Scholar]
- 33. Winokur P. L., et al. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44:2777–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winokur P. L., Vonstein D. L., Hoffman L. J., Uhlenhopp E. K., Doern G. V. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wray C., Davies R. H. 2000. Salmonella infection in cattle, p. 169–190 In Wray C., Wray A. (ed.), Salmonella in domestic animals. CABI Publishing, Wallinford, United Kingdom [Google Scholar]
- 36. Zioga A., et al. 2008. Evidence for chromosomal and plasmid location of CMY-2 cephalosporinase gene in Salmonella serotype Typhimurium. J. Antimicrob. Chemother. 61:1389–1390 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.