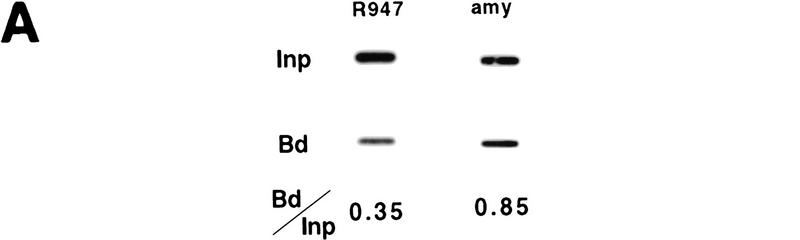
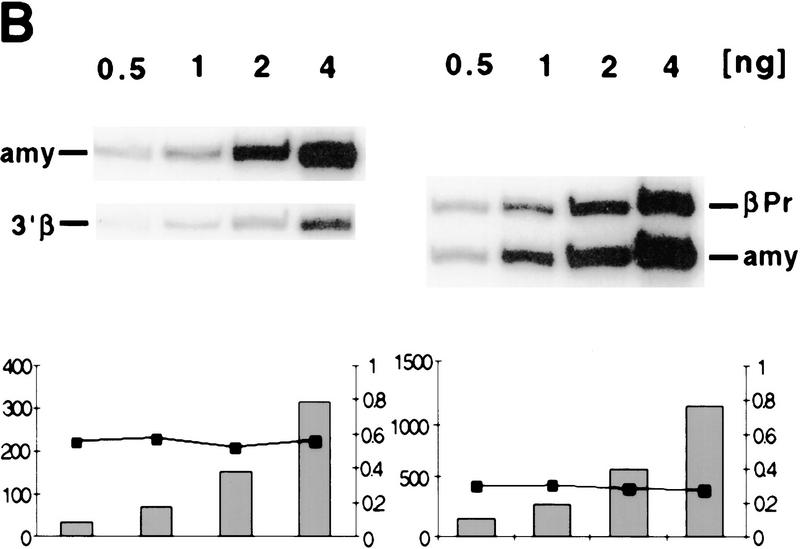
Figure 2.
Immunoprecipitation and duplex PCR assays. (A) Depletion of centromeric (heterochromatic) sequences and an inactive mouse gene in chromatin enriched for acetylated histone H4. Chromatin from MEL cells was immunoprecipitated with an antibody that detects all acetylated isoforms of histone H4 (αH4–Ac). Input and antibody-bound DNA (500 ng) were slot blotted and hybridized with an oligomer corresponding to a murine centromeric minor satellite repeat (R947, Kipling et al. 1994). This sequence is 2.8-fold less abundant in the antibody bound fraction. The same blot was rehybridized with a probe from the mouse amylase gene (generated by PCR with the primer pairs amy4 and amy6, see Materials and Methods). This pancreatic-specific gene, which is inactive in a red cell background, is slightly less abundant in chromatin enriched for acetylated H4. (B) Abundance of human and mouse globin sequences in chromatin enriched for acetylated histones was determined relative to the mouse amylase gene using a duplex PCR assay (see text and Fig. 3). One primer pair amplifies a sequence from the mouse amylase gene, the other pair amplifies either a human or mouse β-globin locus sequence. To determine conditions of linear amplification, serial dilutions of chromatin containing 0.5–4 ng of DNA were used as template. Shown are products and quantification for two representative primer pairs (βPr with amy4 + 6 and 3′β with amy4 + 5), revealing linear amplification of the total signal (bars) and a constant ratio (line) for the two products under these PCR conditions (see Materials and Methods). For the quantitative analysis of immunoprecipitated material shown in Figs. 3 and 4, 1–2 ng of DNA were used per reaction to ensure amplification in the linear range.