Abstract
The availability of oxygen (O2) in aerated (i.e., water-unsaturated) soils affects the metabolic activities of aerobic and anaerobic soil prokaryotes that degrade plant-derived saccharides. Fluctuating availabilities of O2 were imposed on agricultural soil slurries supplemented with cellobiose. Slurries were subjected to oxic conditions (48 h), followed by an anoxic period (120 h) and a final oxic period (24 h). Redox potential was stable at 500 mV during oxic periods but decreased rapidly (within 10 h) under anoxic conditions to −330 mV. The consumption of cellobiose occurred without apparent delay at all redox potentials. The metabolic activities of seven previously identified saccharolytic family-level taxa of the investigated soil were measured with newly designed quantitative PCR assays targeting the 16S rRNA. Four taxa responded to the experimental conditions. The amounts of rRNAs of Micrococcaceae and Cellulomonadaceae (Actinobacteria) increased under oxic conditions. In contrast, the RNA contents of Clostridiaceae (cluster I, Firmicutes) and two uncultured family-level-taxa, i.e., “Cellu” and “Sphingo” (both Bacteroidetes) increased under anoxic conditions. That the degradation of cellobiose was independent of the availability of O2 and that redox potentials decreased in response to anaerobic activities indicated that the degradation of cellobiose was linked to functionally redundant cellobiose-degrading taxa capable of altering redox conditions.
INTRODUCTION
Cellobiose is the main product of microbial hydrolysis of cellulose under both anoxic and oxic conditions in soils (25). Cellulose is the most abundant biopolymer in terrestrial ecosystems. Thus, cellobiose is a major source of carbon for soil bacterial communities (23, 25). Degradation of cellobiose is catalyzed by various aerobic and anaerobic microorganisms (2, 25). The distribution of oxic and anoxic microzones in aerated soils is dynamic, spatially heterogeneous, and dependent on soil moisture and soil aggregate size (11, 30, 49). Agricultural soils (except for flooded rice fields) are usually water unsaturated and aerated. Nonetheless, anoxic microzones, e.g., in water-saturated soil pores, with low concentrations of O2 and low redox potential occur, in which anaerobic microbial activities can take place (7, 22, 32, 34). Alternative electron acceptors, such as nitrate, manganese oxide, or ferric iron, are utilized in anoxic zones, and redox potentials may drop to negative values (11, 31). Thus, fluctuation of O2 availability is an environmental factor that affects the metabolic activities of specific bacterial taxa in water-unsaturated soils (20, 30).
Functional redundancy in a microbial biome (i.e., the concomitant occurrence of different microbial taxa that have the capacity to utilize the same substrate) might be important to stable biome function under fluctuating environmental conditions. For example, although glucose consumption rates in aerated soil can vary in response to redox potential, the overall capacity of the microbial biome to consume glucose is independent of redox potential (34, 39). Also, the availability of O2 does not appreciably affect nitrogen turnover rates in tropical forest soil, even though the presence or absence of O2 engages different members of the microbial community that are involved in nitrogen turnover (32, 33). Although previous studies have evaluated the aerobic and anaerobic activities of saccharolytic microorganisms of agricultural soils (3, 18, 40), how functionally redundant taxa respond to periodic changes of the availability of O2 has not been investigated. The objectives of this study were to (i) determine if functionally redundant cellobiose-utilizing taxa in an aerated soil rapidly respond to the fluctuating availabilities of O2, (ii) resolve how these taxa affect redox potentials, and (iii) evaluate the bacterial constituents of the microbial biome that responded to cellobiose under these contrasting conditions.
MATERIALS AND METHODS
Sampling site and soil characteristics.
Soil from the upper 20 cm of a wheat-planted agricultural soil was sampled in April 2010. The study site is located near Munich (Germany) on the research farm Klostergut Scheyern (48°30.0′N, 11°20.7′E). The soil pH was 6.9, and the water content was 13.8%. Ammonium, and nitrate concentrations were 0.2 ± 0.0 and 1.3 ± 0.1 mg (g soil dry weight)−1, respectively. Total amounts of iron, manganese, and sulfate were 32.5 ± 0.3, 0.9 ± 0.0, and 0.04 ± 0.0 mg (g soil dry weight)−1, respectively. Ferrous iron was not detectable. Soil samples were stored at 2°C and sieved (mesh size, 2 mm) before use.
Soil slurries.
Soil slurries were prepared in triplicates in cylindrical Plexiglas chambers (1,000 ml) that could be permanently flushed with gas (see Fig. S1 in the supplemental material). Fifty grams of fresh, sieved soil was filled in a chamber, 250 ml sterile oxic or anoxic water was added, chambers were closed, and the slurries were mixed thoroughly by manual shaking. Chambers were flushed with sterile N2 (100%; Rießner, Germany; 3 replicates) or with synthetic air (80% N2, 20% O2; Rießner, Germany; 6 replicates) for 1 h. The chambers were then divided into three groups: anoxic controls (permanently anoxic, no substrate supplementation), redox controls (changing O2 status, no substrate supplementation), and cellobiose-supplemented slurries (changing O2 status). Cellobiose was pulsed four times, i.e., after 0, 48, 96, and 168 h. Total amounts of cellobiose (0.15 mmol substrate per pulse) were 0.6 mmol, which corresponds to 7.2 mmol of carbon. After 48 h, slurries were flushed with N2 for 1.5 h to achieve anoxic conditions. Reaeration was performed after 168 h in redox controls and substrate-supplemented treatments by constant flushing with synthetic air until the end of incubation. A duration of 168 h for anoxic incubation was used since similar periods of O2 depletion are likely to occur in upland soils (32). Chambers were incubated at 20°C. Incubation was done static, but slurries were mixed manually every 1 to 2 h for redox measurements and sampling. Liquid samples were taken with sterile syringes and either directly analyzed or stored at −80°C.
Chemical analyses.
The pH was measured with a U457-S7/110 combination pH electrode (Ingold, Germany). The redox potential was measured with an EMC 30 L electrode (Sensortechnik Meinsberg, Germany). Soil moisture content was determined by quantification of weight loss of fresh soil after drying at 105°C for 48 h (41). Ferrous iron was determined photometrically (43). Total concentrations of nitrate, iron, manganese, sulfate, and ammonium were analyzed by ion chromatography (Central Analytics, University of Bayreuth, Germany). Nitrate, iron, manganese, and sulfate were extracted by acidic hydrolysis (HF-HNO3-HCl; 1:3:2). Ammonium concentrations were analyzed in the aqueous phase. Dissolved organic compounds (detection limit, ≥50 μM) were measured by high-performance liquid chromatography (HPLC) as previously published (10, 27). Gaseous compounds were not measured, since the gas phase was periodically exchanged.
Extraction of nucleic acids.
Nucleic acids were extracted from treatments exposed to fluctuating availabilities of O2 after 0, 48, 102, 168, and 196 h from 2 ml of soil slurry. A protocol including bead beating and phenol-chloroform extraction was used (17). DNA and RNA were obtained by RNase A and DNase I (Fermentas GmbH, Germany) treatment of aliquoted nucleic acids, respectively. Nucleic acids were precipitated with 0.7 volume of isopropanol (100%) and 0.1 volume of sodium chloride (5 M) overnight at −20°C. Pellets were washed with ice-cold ethanol (70%) and resuspended in diethyl pyrocarbonate-treated water. DNA and RNA concentrations were quantified with the Quant-iT PicoGreen RNA assay kit and Quant-iT RiboGreen assay kit (Invitrogen, Germany), respectively. RNA samples were used as controls in a subsequent 16S rRNA gene PCR but did not yield a PCR product, indicating that the RNA was DNA free. Nucleic acid solutions were stored at −80°C.
Quantification of 16S rRNA genes and transcripts.
Reverse transcription of RNA was performed with random hexamer primers and SuperScript III first-strand synthesis supermix (Invitrogen, Germany) according to the manufacturer's protocol. Primers and protocols used for quantitative PCR (qPCR) are shown in Table S1 in the supplemental material. All environmental DNA and cDNA samples were diluted 20- or 10-fold, respectively, to reduce potential inhibition of qPCR by coextracted PCR-inhibiting compounds. Five microliters of DNA or cDNA was used as the template in 20-μl reaction mixtures containing 1-fold SensiMix (Bioline, Germany), 500 to 1,500 nM each primer (see Table S1 in the supplemental material), and sterilized deionized water. Initial denaturation was at 94°C for 8 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at temperatures appropriate for each primer set (see Table S1 in the supplemental material) for 30 s, and elongation at 72°C for 40s. The fluorescence signal was recorded during elongation. Final elongation was at 72°C for 5 min. Melting curve analyses were performed from 70 to 94°C with increments of 0.15°C per cycle. Agarose gel electrophoreses revealed that any qPCR assay gained a single band of the expected length (see Table S1 in the supplemental material), suggesting that unspecific PCR products (i.e., amplicons shorter or longer than expected) did not form (data not shown).
Family-level assays (see Table S1 in the supplemental material) did not yield sufficient gene amplification in DNA samples (i.e., target sequences were below the detection limit of about 10 genes per reaction; threshold cycle [CT], >30). The results refer exclusively to transcript numbers quantified in cDNA samples. Transcript numbers were corrected for inhibition by measuring inhibition factors for all samples as described previously (47). The total amount of RNA that was extracted from experimental replicates was variable (see Fig. S2 in the supplemental material). Thus, 16S rRNA transcript numbers were quantified per ng RNA rather than per gram (dry weight) of soil. These absolute numbers of transcripts of family-level taxa were divided by the corresponding number of transcripts of total bacteria to further correct for variability in lysis efficiencies and the growth of bacteria. For each replicate, these copy numbers of family-level taxa were normalized based on the highest copy number measured (i.e., the highest number obtained for each assay, including cellobiose-supplemented and unsupplemented soil slurries, was set at 100%). Data presented are means of experimental triplicates (including standard deviations) or duplicates (without standard deviation).
Target group specificity of qPCR assays.
qPCR products from each assay of every slurry sample (i.e., all replicates and time points) were pooled and sent to LGC Genomics (Berlin, Germany) for cloning and sequencing. Phylogenetic analyses were performed with the Mega 4 software (http://www.megasoftware.net/) (21, 44) and the ARB software package (http://www.arb-home.de; version 2005) (24) using the latest database release of SILVA (release 104) (35). The specificities of the qPCR assays are shown in Table 1.
Table 1.
Specificity of qPCR assays
| Assay(s) | % (no. of sequences)a |
Affiliation of nontargets (no. of sequences) | ||
|---|---|---|---|---|
| Targets | Nontargets | |||
| Family level | ||||
| Micrococcaceae and Cellulomonadaceae | 100.0 (19) | 0.0 (0) | ||
| Kineosporiaceae and Nocardioidaceae | 78.9 (15) | 21.1 (4) | Acidobacteria (1), Cyanobacteria (2), Firmicutes (1) | |
| Cluster I Clostridiaceae | 84.2 (16) | 15.8 (3) | Gammaproteobacteria (3) | |
| Planctomycetaceae | 100.0 (20) | 0.0 (0) | ||
| “Cellu”b (Bacteroidetes) | 95.5 (21) | 4.5 (1) | Cyanobacteria (1) | |
| “Sphingo”b (Bacteroidetes) | 61.1 (11) | 38.9 (7) | Deltaproteobacteria (3), Gemmatimonadetes (4) | |
| “Deha”b (Chloroflexi) | 80.0 (16) | 20.0 (4) | Nitrospirae (2), Firmicutes (2) | |
| Domain level | ||||
| Bacteria | 100.0 (20) | 0.0 (0) | ||
Parenthetical values are the number of sequences that were obtained by cloning and sequencing of qPCR products amplified with the specific primer pair (see Table S1 in the supplemental material) in the corresponding assay.
Name and affiliation as defined elsewhere (40).
Statistical analyses.
Dependent t tests (one-way analysis of variance [ANOVA]) were used to analyze pairwise whether the mean copy numbers for two time points within one specific assay were statistically different. Variables used were as follows: n = 3 to 6; degree of freedom fd = −1; significance level α = 0.05; variance ν = 2.9200 (n = 3) or 2.0150 (n = 6). t > ν implies a statistically significant difference between a data point and its preceding value.
Nucleotide sequence accession numbers.
Sequences were deposited at EMBL under accession numbers FR773527 to FR773699.
RESULTS
Effect of cellobiose on redox potentials.
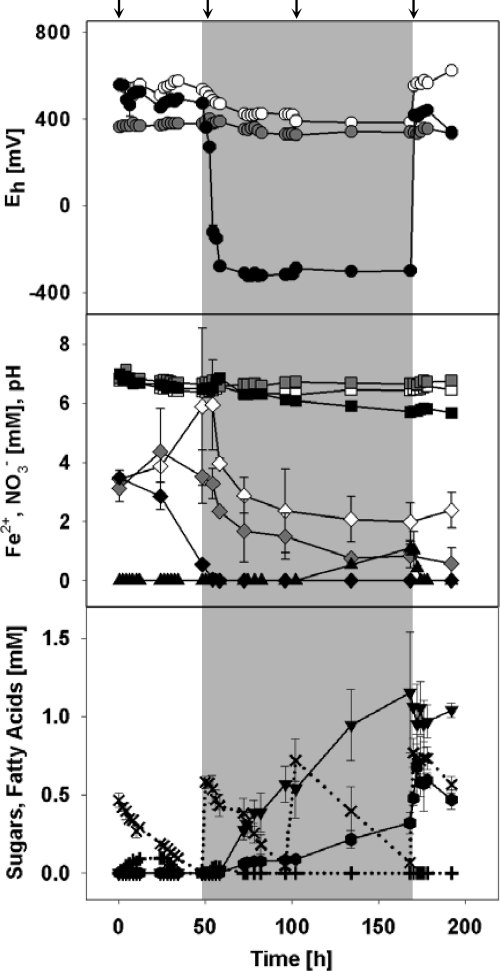
The initial redox potentials in oxic and anoxic slurries were 512 ± 1 mV and 363 ± 3 mV, respectively (Fig. 1). The redox potentials of anoxic unsupplemented slurries were relatively stable and decreased only slightly from the initial values to 348 ± 12 mV (Fig. 1). The redox potentials of cellobiose-supplemented slurries decreased rapidly during the anoxic period to −299 ± 6 mV and remained constant until reaeration (Fig. 1). Reaeration increased redox potentials within 2 to 4 h to 573 ± 25 mV in unsupplemented slurries and to 411 ± 35 mV in cellobiose-supplemented slurries (Fig. 1).
Fig. 1.
Substrate and product concentrations, redox potentials, and pHs of cellobiose treatments, including control treatments. Closed symbols are values obtained from cellobiose-supplemented treatments, empty symbols are values obtained from unsupplemented treatments with changing O2 availability, and gray symbols are values obtained from unsupplemented permanently anoxic treatments. The shaded areas correspond to the anoxic period. Symbols: ●, Eh; ▪, pH; ♦, nitrate; ▴, iron(II); ✖, cellobiose (dotted line); ✚, glucose (dotted line); ▾, acetate;  , lactate. The arrows indicate when substrates were added.
, lactate. The arrows indicate when substrates were added.
Cellobiose consumption and product formation.
Supplemented cellobiose was consumed without apparent delay during both oxic and anoxic periods (i.e., the mean consumption rate was 11 ± 2 μmol cellobiose h−1 liter−1), indicating that the microbial community was poised to both aerobically and anaerobically consume this substrate (Fig. 1). The first and second pulses of cellobiose resulted in a transient accumulation of small amounts of glucose, whereas no glucose was detected at later time points, indicating that active microbes increased in cell numbers and/or consumed cellobiose more efficiently (Fig. 1). Nitrate concentrations decreased rapidly during the first oxic phase in cellobiose-supplemented treatments (Fig. 1), suggesting that nitrate was reductively assimilated. Several fermentation products and ferrous iron were formed after redox potentials decreased to values below −300 mV (Fig. 1). Formate and succinate were produced in small amounts (approximately 0.25 mM) during the first part of the anoxic phase, but formate was subsequently consumed during the later part of the anoxic phase (data not shown). Acetate and lactate accumulated up to 1 mM during the anoxic phase (Fig. 1). Acetate, succinate, and ferrous iron, but not lactate, decreased after reaeration, indicating that they were dissimilated, assimilated, or chemically oxidized. In unsupplemented slurries, nitrate concentrations increased during oxic phases but slowly decreased during the anoxic phase (Fig. 1), indicating that nitrate was formed by nitrification in response to O2 and subsequently consumed anaerobically in the absence of O2. A linear decrease in the concentration of nitrate was detected in anoxic controls (Fig. 1). The pH decreased from 6.6 ± 0.2 to 5.7 ± 0.0 in cellobiose-supplemented slurries but was stable at approximately 6.6 in unsupplemented control treatments (Fig. 1).
Response of bacterial taxa to O2 availability.
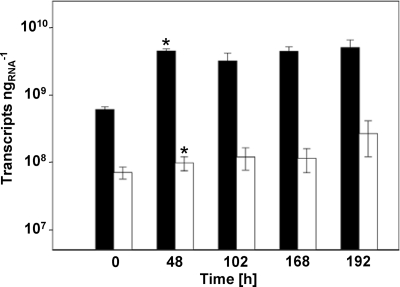
The total amount of 16S rRNA of bacteria significantly increased from 0.6 × 109 to 5.0 × 109 transcripts ng RNA−1 during the first oxic period (0 to 48 h) (Fig. 2), but neither the shift from oxic to anoxic conditions (48 to 168 h) nor reaeration (168 to 192 h) altered total bacterial 16S rRNA molecule numbers (Fig. 2). A similar trend was also observed in controls without cellobiose (Fig. 2). Thus, an increased availability of soil endogenous carbon compounds due to the experimental conditions likely stimulated soil bacteria.
Fig. 2.
Numbers of 16S rRNA gene transcripts of Bacteria in soil slurries. The filled bars are values obtained from cellobiose treatments. The empty bars are values obtained from unsupplemented control treatments with changing availability of O2. An asterisk indicates that a transcript number is statistically different from the transcript number of the previous time point (t test).
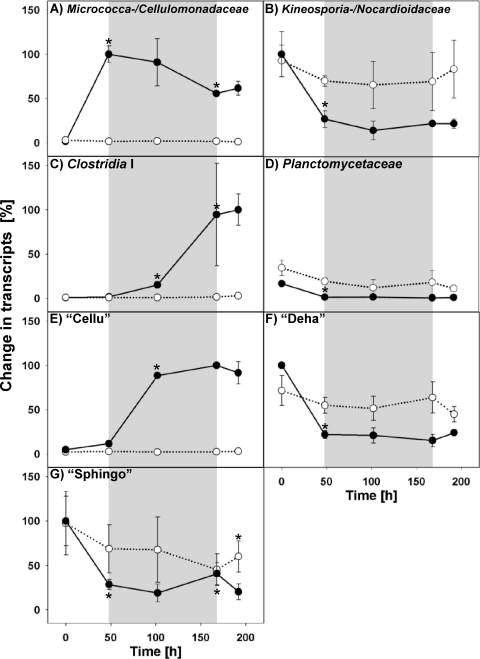
The Bacteria-targeting primers were highly specific and did not yield any nontarget taxa. 16S rRNA gene transcript numbers of Micrococcaceae/Cellulomonadaceae increased in the first oxic period (Fig. 3A), leading to highest 16S rRNA numbers of all measured bacterial taxa (see Fig. S3 in the supplemental material). In contrast, transcript numbers of Clostridia I and the two novel family-level taxa “Cellu” and “Sphingo” (both Bacteroidetes) (40) increased under anoxic conditions (48 to 168 h) in cellobiose-supplemented slurries (Fig. 3C and E). A stimulation of Kineosporiaceae/Nocardioidaceae, Planctomycetaceae, and the uncultured taxon “Deha” (Chloroflexi) (40) by cellobiose supplementation was not apparent (Fig. 3B, D and F). None of the assessed family level-taxa was stimulated in unsupplemented slurries (Fig. 3).
Fig. 3.
Effects of cellobiose on taxon-specific 16S rRNA gene transcripts. The shaded areas correspond to the anoxic period. Closed symbols are values obtained from cellobiose treatments, and empty symbols are values obtained from unsupplemented control treatments. Values are the means for two replicates (without standard deviation) or three replicates (with standard deviation). The highest number of taxon-specific transcripts per assay (i.e., cellobiose and unsupplemented control treatments) was set at 100% (reference values are shown in Table S2 in the supplemental material; absolute values are shown in Table S3 in the supplemental material). An asterisk indicates that a transcript number is statistically different from the transcript number of the previous time point (t test).
DISCUSSION
The redox potential of soil is determined by various factors, the most important ones being the availabilities of O2 and organic compounds for soil microbial biomes (5, 7, 12). High aerobic activities of soil microorganisms can be coupled to a rapid depletion of O2 and a rapid change in redox conditions (7, 11). Nevertheless, a biome function, e.g., the capacity to degrade a certain saccharide, may remain stable under changing redox conditions (32, 33). In this regard, the capacity of the biome of the soil to readily consume supplemental cellobiose was not affected by the availability of O2 (Fig. 1).
Effects of degradation of cellobiose on the redox potential of soil slurries.
A rapid decrease of the redox potential in cellobiose-supplemented treatments occurred when O2 was removed; i.e., the redox potential decreased more than 700 mV within 10 h (Fig. 1). It is likely that this change in redox potential was due to the metabolic activity of cellobiose-utilizing microbes (i.e., the oxidation of cellobiose coupled to the reduction of high-redox-potential electron acceptors [e.g., nitrate and ferric iron]). The formation of nitrate in unsupplemented slurries under oxic conditions indicated that nitrate formation by nitrification exceeded nitrate consumption (Fig. 1). Most fermentations and the reduction of ferric iron are thermodynamically unfavorable at such a high redox potential during the oxic period (5). The reduction of nitrate and iron in water-unsaturated bulk soil can occur at 230 mV and 150 mV, respectively (26). The presence of nitrate might have been responsible for (i) the stabilization of redox potentials above 320 mV during the anoxic period in unsupplemented and in the permanently anoxic control slurries (Fig. 1) and (ii) the apparent lack of ferrous iron and fermentation products (11, 34).
However, nitrate was completely consumed during the first 50 h in cellobiose-supplemented slurries, and this was followed by the apparent reduction of ferric iron (Fig. 1), suggesting that nitrate-utilizing microorganisms were highly active. The rapid decrease of the redox potential in these slurries was likely due to the large amounts of readily accessible organic carbon (i.e., cellobiose) that led to the rapid reduction of electron acceptors that have positive redox potentials (11, 34). Products indicative of different fermentations (e.g., mixed acid and/or lactic acid fermentations [16]) accumulated during the anoxic period (Fig. 1). The fermentation products were similar to those detected during the anaerobic degradation of sugars by various soils (11, 13, 22). Certain fermentations occur optimally at redox potentials of below −100 mV (11, 34), conditions that were achieved in cellobiose-supplemented soil slurries during the anoxic period. Reaeration of cellobiose-supplemented slurries resulted in a strong increase of the redox potential and rapid consumption of organic acids and ferrous iron (Fig. 1), the latter processes likely being mediated by heterotrophic aerobes. Classical lactate fermentation can occur in the presence of O2 and is catalyzed by aerotolerant anaerobes (6). Hence, although such aerotolerant soil lactate fermenters (6, 28) were not targeted by qPCR, the accumulation of lactate in cellobiose treatments and the known occurrence of these taxa in soils (8, 28) suggest that lactate fermenters were active during both oxic and anoxic phases.
Correlations between metabolic activities and taxa.
Micrococcaceae and Cellulomonadaceae (Actinobacteria), families that are numerically important in soils and that contain aerobic cellulolytic and saccharolytic genera (19, 42), were the taxa that responded most pronouncedly to cellobiose supplementation under oxic conditions (Fig. 3; see Fig. S3 in the supplemental material). Both families have previously been identified as important taxa involved in the degradation of cellobiose in the investigated agricultural soil (40). The majority of the amplified gene sequences were closely related to the genera Arthrobacter and Cellulomonas. These genera contain aerobic isolates that can grow on cellobiose (1, 46). Although certain Arthrobacter and Cellulomonas species are capable of anaerobic growth by fermentation, most known species of these genera are strict aerobes or grow best aerobically (1, 15, 42). The primary aerobic phenotype of known isolates of these taxa fits well with their apparent repression during the anoxic period (Fig. 3A).
Cluster I Clostridiaceae (Firmicutes) and two novel family-level taxa (i.e., “Cellu” and “Sphingo”) of the phylum Bacteroidetes (40) were stimulated by cellobiose supplementation exclusively under anoxic conditions (Fig. 3C, E, and G). Sequences that were amplified with the Clostridiaceae-specific qPCR assay were closely related to members of Clostridium, a taxon that contains saccharolytic anaerobes (e.g., Clostridium puniceum, with 98% similarity to sequences obtained in this study [9, 48]). Hence, the detection of Clostridium-affiliated sequences in anoxic treatments was consistent with known strict anaerobic clostridial phenotypes. Genotypes retrieved from the “Cellu”-specific qPCR assay were not affiliated with cultured species (i.e., they were more than 11% dissimilar to 16S rRNAs of cultured members). Thus, the detected members of “Cellu” likely represent a novel phylogenetically deep-branching group of saccharolytic anaerobes. The “Sphingo” qPCR assay detected two nontarget taxa, i.e., Gemmatimonadaceae (Gemmatimonadetes) and Polyangiaceae (Deltaproteobacteria). That the nontarget families include only strictly aerobic species (36, 50) suggests that the increase in “Sphingo”-affiliated rRNA was caused by species of Sphingobacteriales that were active under anoxic conditions. The capacity to utilize cellulose and cellulose-derived saccharides (e.g., cellobiose) is common to various families of Bacteroidetes (e.g., Bacteroidaceae, Cytophagaceae, and Flavobacteriaceae [4, 29, 37, 38]). Most of the detected Bacteroidetes genotypes in the current study were affiliated with Cytophaga and Flexibacter (Cytophagaceae). Species of Cytophaga are cellulolytic and widely distributed in terrestrial environments (36). Flexibacter species are important fish pathogens, and a role for this genus in terrestrial habitats is unknown (36).
Members of Kineosporiaceae/Nocardioidaceae (Actinobacteria), Planctomycetaceae (Planctomycetes), and the novel family-level taxon “Deha” (Chloroflexi) incorporated 13C derived from supplemented 13C-labeled sugars in a previous experiment on the same soil (40). Although these families were part of the active cellulolytic and saccharolytic community in a previous study and might be well adapted to low concentrations of cellulose-derived sugars under certain conditions (40), they were apparently out competed or did not respond remarkably during the short incubation time of the experiment (Fig. 3B, D, and F).
Conclusions and future perspectives.
The availability of O2 may change rapidly in aerated soils (14, 26, 45). The bacterial utilization of supplemental cellobiose resulted in a rapid decrease of the redox potential when O2 was no longer available. There was no apparent delay in the consumption of cellobiose during anoxic or oxic periods (Fig. 1). The results of the current study (i) suggest that bacterial taxa that are involved in the degradation of plant-derived saccharides can respond rapidly to differing availabilities of O2 and alter redox conditions and (ii) confirm that the differential engagement of functionally redundant taxa with different phenotypic adaptations to a fluctuating environmental factor (e.g., availability of O2) contributes to the stability of a particular function of the soil microbial biome (e.g., cellobiose degradation). Determining how other environmental factors, e.g., anthropogenic factors, might affect functionally redundant bacterial taxa is a topic that warrants further study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stephan Schulz, Michael Schloter, and Jean Charles Munch for providing soil samples. We are grateful to Gerhard Küfner, Peter Schmidt, and colleagues for assistance in the planning and construction of the incubation chambers.
Support for this study was provided by the Deutsche Forschungsgemeinschaft (grant Ko2912/3-1, DFG Priority Program 1315 “Biogeochemical Interfaces in Soil”) and the University of Bayreuth.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Baganara C., Toci R., Gaudin C., Belaich J. P. 1985. Isolation and characterization of a cellulolytic microorganism, Cellulomonas fermentans sp. nov. Int. J. Syst. Bacteriol. 35:502–507 [Google Scholar]
- 2. Baldrian P., Valaskova V. 2008. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 32:501–521 [DOI] [PubMed] [Google Scholar]
- 3. Bernard L., et al. 2007. Dynamics and identification of soil microbial populations actively assimilating carbon from 13C-labelled wheat residue as estimated by DNA- and RNA-SIP techniques. Environ. Microbiol. 9:752–764 [DOI] [PubMed] [Google Scholar]
- 4. Bernardet J. F., et al. 1996. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom.nov. (Basonym, Cytophaga aquatilis Strohl and Tait 1978). Int. J. Syst. Bacteriol. 46:128–148 [Google Scholar]
- 5. Bohn H. L. 1971. Redox potentials. Soil Sci. 112:39–45 [Google Scholar]
- 6. Brioukhanov A. L., Netrusov A. I. 2007. Aerotolerance of strictly anaerobic microorganisms and factors of defense against oxidative stress: a review. Appl. Biochem. Microbiol. 43:567–582 [PubMed] [Google Scholar]
- 7. Brune A., Frenzel P., Cypionka H. 2000. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol. Rev. 24:691–710 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y. S., Yanagida F., Shinohara T. 2005. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett. Appl. Microbiol. 40:195–200 [DOI] [PubMed] [Google Scholar]
- 9. Collins M. D., et al. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812–826 [DOI] [PubMed] [Google Scholar]
- 10. Daniel S. L., Hsu T., Dean S. I., Drake H. L. 1990. Characterization of the H2-dependent and CO-dependent chemolithotrophic potentials of the acetogens Clostridium thermoaceticum and Acetogenium kivui. J. Bacteriol. 172:4464–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dassonville F., Renault P., Valles V. 2004. A model describing the interactions between anaerobic microbiology and geochemistry in a soil amended with glucose and nitrate. Eur. J. Soil Sci. 55:29–45 [Google Scholar]
- 12. de Boer W., Folman L. B., Summerbell R. C., Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29:795–811 [DOI] [PubMed] [Google Scholar]
- 13. Degelmann D., Kolb S., Dumont M., Murrell J. C., Drake H. L. 2009. Enterobacteriaceae facilitate the anaerobic degradation of glucose by a forest soil. FEMS Microbiol. Ecol. 68:312–319 [DOI] [PubMed] [Google Scholar]
- 14. Denef K., et al. 2001. Influence of dry-wet cycles on the interrelationship between aggregate, particulate organic matter, and microbial community dynamics. Soil Biol. Biochem. 33:1599–1611 [Google Scholar]
- 15. Eschbach M., Möbitz H., Rompf A., Jahn D. 2003. Members of the genus Arthrobacter grow anaerobically using nitrate ammonification and fermentative processes: anaerobic adaptation of aerobic bacteria abundant in soil. FEMS Microbiol. Lett. 223:227–230 [DOI] [PubMed] [Google Scholar]
- 16. Gottschalk G. 1989. Bacterial fermentations, p. 210–282.In Gottschalk G. (ed.), Bacterial metabolism, 2nd ed. Springer, New York, NY [Google Scholar]
- 17. Griffiths R. I., Whiteley A. S., O'Donnell A. G., Bailey M. J. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haichar F. E. Z., et al. 2007. Identification of cellulolytic bacteria in soil by stable isotope probing. Environ. Microbiol. 9:625–634 [DOI] [PubMed] [Google Scholar]
- 19. Jones D., Keddie R. M. 2006. The genus Arthrobacter, p. 945–960.In Dworkin M. M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. (ed.), The prokaryotes. Springer, New York, NY [Google Scholar]
- 20. Kremen A., Bear J., Shavit U., Shaviv A. 2005. Model demonstrating the potential for coupled nitrification denitrification in soil aggregates. Environ. Sci. Technol. 39:4180–4188 [DOI] [PubMed] [Google Scholar]
- 21. Kumar S., Dudley J., Nei M., Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Küsel K., Drake H. L. 1995. Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl. Environ. Microbiol. 61:3667–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li T., Mazeas L., Sghir A., Leblon G., Bouchez T. 2009. Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ. Microbiol. 11:889–904 [DOI] [PubMed] [Google Scholar]
- 24. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lynd L. R., Weimer P. J., van Zyl W. H., Pretorius I. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mansfeldt T. 2004. Redox potential of bulk soil and soil solution concentration of nitrate, manganese, iron and sulfate in two Gleysols. J. Plant Nutr. Soil Sci. 167:7–16 [Google Scholar]
- 27. Matthies C., Freiberger A., Drake H. L. 1993. Fumarate dissimilation and differential reductant flow by Clostridium formicoaceticum and Clostridium aceticum. Arch. Microbiol. 160:273–278 [Google Scholar]
- 28. Matthies C., Gössner A., Acker G., Schramm A., Drake H. L. 2004. Lactovum miscens gen. nov., sp. nov., an aerotolerant, psychrotolerant, mixed-fermentative anaerobe from acidic soil. Res. Microbiol. 155:847–854 [DOI] [PubMed] [Google Scholar]
- 29. Nakagawa Y., Yamasato K. 1996. Emendation of the genus Cytophaga and transfer of Cytophaga agarovorans and Cytophaga salmonicolor to Marinilabilia gen. nov.: phylogenetic analysis of the Flavobacterium-Cytophaga complex. Int. J. Syst. Bacteriol. 46:599–603 [Google Scholar]
- 30. Or D., Smets B. F., Wraith J. M., Dechesne A., Friedman S. P. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media—a review. Adv. Water Resour. 30:1505–1527 [Google Scholar]
- 31. Peters V., Conrad R. 1996. Sequential reduction processes and initiation of CH4 production upon flooding of oxic upland soils. Soil Biol. Biochem. 28:371–382 [Google Scholar]
- 32. Pett-Ridge J., Firestone M. K. 2005. Redox fluctuation structures microbial communities in a wet tropical soil. Appl. Environ. Microbiol. 71:6998–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pett-Ridge J., Silver W. L., Firestone M. K. 2006. Redox fluctuations frame microbial community impacts on N-cycling rates in a humid tropical forest soil. Biogeochemistry 81:95–110 [Google Scholar]
- 34. Picek T., Simek M., Santruckova H. 2000. Microbial response to fluctuation of soil aeration status and redox conditions. Biol. Fertil. Soils 31:315–322 [Google Scholar]
- 35. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned rRNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reichenbach H. 2005. Family III. Polyangiaceae, p. 1132–1136.In Boone D. R., Castenholz R. W., Garrity G. M. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, Springer, New York, NY. [Google Scholar]
- 37. Reichenbach H. 2006. The order Cytophagales, p. 549–590.In Dworkin M. M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. (ed.), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 38. Robert C., Chassard C., Lawson P. A., Bernalier-Donadille A. 2007. Bacteriodes cellulosilyticus sp. nov., a cellulolytic bacterium form the gut microbial community. Int. J. Syst. Evol. Microbiol. 57:1516–1520 [DOI] [PubMed] [Google Scholar]
- 39. Santruckova H., Picek T., Tykva R., Simek M., Pavlu B. 2004. Short-term partitioning of 14C-[U]-glucose in the soil microbial pool under varied aeration status. Biol. Fertil. Soils 40:386–392 [Google Scholar]
- 40. Schellenberger S., Kolb S., Drake H. L. 2010. Metabolic responses of novel cellulolytic and saccharolytic agricultural soil bacteria to oxygen. Environ. Microbiol. 12:845–861 [DOI] [PubMed] [Google Scholar]
- 41. Schlichting E., Blume H. P., Stahr K. 1995. Bodenkundliches Praktikum. Blackwall Wissenschaftsverlag, Berlin, Germany [Google Scholar]
- 42. Stackebrandt E., Schuhmann P., Prauser H. 2006. The family Cellulomonadaceae, p. 983–1001.In Dworkin M. M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. (ed.), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 43. Tamura H., Goto K., Yotsuyan T., Nagayama M. 1974. Spectrophotometric determination of iron(II) with 1,10-phenanthroline in presence of large amounts of iron(III). Talanta 21:314–318 [DOI] [PubMed] [Google Scholar]
- 44. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 45. Vorenhout M., van der Geest H. G., van Marum D., Wattel K., Eijsackers H. J. P. 2004. Automated and continuous redox potential measurements in soil. J. Environ. Qual. 33:1562–1567 [DOI] [PubMed] [Google Scholar]
- 46. Wauters G., Charlier J., Janssens M., Delmee M. 2000. Identification of Arthrobacter oxydans, Arthrobacter luteolus sp. nov., and Arthrobacter albus sp. nov. isolated from human clinical specimens. J. Clin. Microbiol. 38:2412–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wieczorek A. S., Drake H. L., Kolb S. 2011. Organic acids and ethanol inhibit oxidation of methane by mire methanotrophs. FEMS Microbiol. Ecol. 77:28–39 [DOI] [PubMed] [Google Scholar]
- 48. Wiegel J. 2009. Family I. Clostridiaceae, p. 736–738.In Boone D. R., Castenholz R. W., Garrity G. M. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 3 Springer, New York, NY. [Google Scholar]
- 49. Zausig J., Stepniewski W., Horn R. 1993. Oxygen concentration and redox potential gradients in unsaturated model soil aggregates. Soil Sci. Soc. Am. J. 57:908–916 [Google Scholar]
- 50. Zhang H., et al. 2003. Gemmatimonas aurantica gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating microorganism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53:1155–1163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.