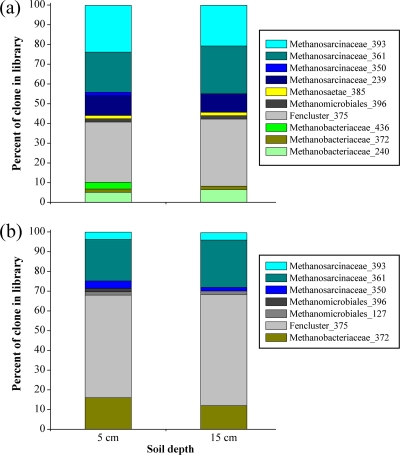
Fig. 4.
Relative proportions of mcrA clones in libraries of the Ermatingen (a) and Heiteren (b) samples from two soil depths (5 and 15 cm) after 6 months of traffic. Only mcrA gene fragments from severely compacted wheel tracks were subjected to cloning and sequencing. Sau96I was used as a restriction enzyme to screen mcrA clones with T-RFLP analyses. T-RFs of 239, 350, 361, and 393 bp are representative of Methanosarcinaceae; T-RFs of 127 and 396 bp are representative of Methanomicrobiales; a T-RF of 375 bp is representative of the Fencluster within Methanomicrobiales; T-RFs of 240, 372, and 436 bp are representative of Methanobacteriaceae; and a T-RF of 385 bp is representative of Methanosaetae.