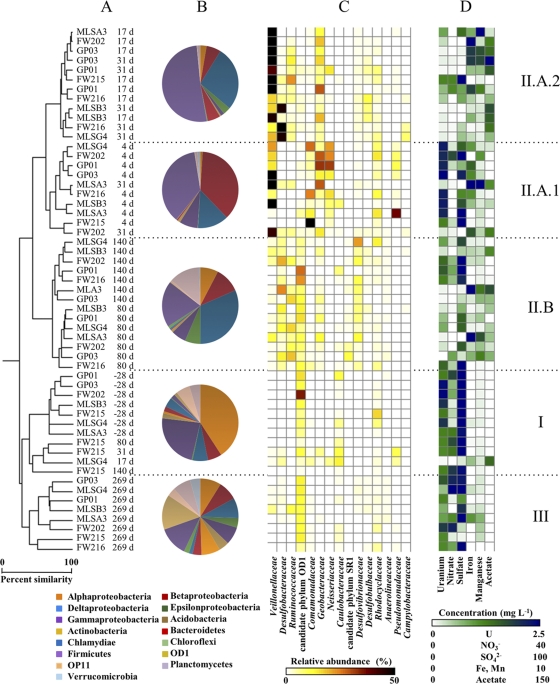
Fig. 2.
Comparisons of groundwater bacterial community structures and composition during the EVO experiment. Sample names indicate the monitoring well identifier (ID) and the sampling time point relative to EVO amendment. (A) Samples clustered according to abundance-based Sorenson's indices of 16S rRNA gene libraries normalized to 2,500 reads each. d, days. (B) Pie charts indicate the mean relative abundance of 16S rRNA gene pyrosequencing reads, classified at the division or class level, for samples corresponding to the indicated clades. (C) The heat map indicates the sum of the relative abundance of those 16S rRNA gene reads classified as members of the indicated taxonomic groups. Relative abundances for the 15 most abundant families or candidate phyla are shown for each sample. (D) The heat map indicates the concentrations of various dissolved constituents in groundwater samples collected at the same time as the microbiological samples.