Abstract
Clostridium perfringensis a Gram-positive anaerobic spore-forming bacterium that is widespread in environmental soil and sewage, as well as in animal intestines. It is also a causative agent of diseases in humans and other animals, and it produces numerous extracellular enzymes and toxins. Although these toxins have been characterized in detail, regulators of toxin genes are less well understood. The present study identified CPE1447 and CPE1446 as novel regulators of toxin gene expression. CPE1447 and CPE1446 are cotranscribed as an operon, and the encoded proteins have a helix-turn-helix (HTH) motif at the N termini of their amino acid sequences, suggesting that CPE1447 and CPE1446 control the target genes as transcriptional regulators. The expression of several genes encoding toxins was changed in both a CPE1446 mutant and a CPE1447-CPE1446 deletion mutant. Complementation of CPE1446 and CPE1447 revealed that CPE1447 and CPE1446 coordinately regulate their target genes. CPE1447 protein was coprecipitated with His-tagged CPE1446 protein, indicating that the CPE1447 and CPE1446 proteins form a stable complex in C. perfringensunder their native conditions. Although the small RNA that regulates several genes under the VirR/VirS two-component system (VR-RNA) positively affected CPE1447-CPE1446 mRNA expression, it did not control expression of the CPE1447-CPE1446 regulon, demonstrating that CPE1447 and CPE1446 regulate a different set of toxin genes from the VirR/VirS–VR-RNA cascade.
INTRODUCTION
The Gram-positive anaerobic spore-forming bacterium, Clostridium perfringensis widespread in the environment and in the gastrointestinal tract. It is pathogenic to humans and other animals and is a causative agent of gas gangrene and food poisoning (20, 21). C. perfringensstrain 13 is an enterotoxin-negative type A strain that produces several toxins, including phospholipase C (alpha-toxin), perfringolysin O (θ-toxin), collagenase (κ-toxin), sialidase, and hyaluronidase (μ-toxin). These toxins are encoded by the genes plc, pfoA, colA, nanI, nanJ, and nagHIJKL, respectively, which are expressed during the exponential growth phase (1, 19). Because C. perfringenshas few genes involved in amino acid biosynthesis, host cell degradation is required to generate nutritional sources, and the expression of genes encoding extracellular enzymes and toxins should be tightly controlled in concert (22).
The primary toxin for gas gangrene, a lethal disease caused by the C. perfringenstype A strain, is phospholipase C, since the plcmutant of C. perfringenscannot cause clostridial myonecrosis in mice, indicating that the alpha-toxin is essential for virulence (2). In contrast, the θ-toxin might play a role in pathogenesis, but it is not essential, since the pfoAmutant can still cause clostridial myonecrosis in mice (2). Sialidase is thought to be associated with the virulence of several bacterial pathogens, such as Vibrio cholerae(11) and Pseudomonas aeruginosa(10). The secreted major sialidase of C. perfingens, NanI, might interact with the extracellular environment of the host tissue. Both NanI and NanJ enhance the alpha-toxin-mediated cytotoxic effect, but they are not essential for virulence (7). Hyaluronidases might also play an important role in enabling the spread of the pathogen, as the spread of their toxins throughout host tissues is enhanced by the degradation of hyaluronate, which is a major constituent of the core substance of most connective tissues (14). The genome of C. perfringensstrain 13 harbors five hyaluronidase genes, suggesting that the enzyme facilitates the virulence of the strain. However, the role of hyaluronidase in the pathogenesis of C. perfringensstrain 13 is unknown.
The major regulatory factor that is thought to be involved in the regulation of toxin production at the exponential phase is the VirR/VirS–VR-RNA cascade, which affects the expression of over 100 genes, including those encoding the alpha-, θ-, κ-, and μ-toxins, as well as sialidase (17, 18). Although VR-RNA directly regulates the expression of κ-toxin (colA) via an antiantisense mechanism (16), how it regulates that of sialidase, as well as the alpha-, θ-, and μ-toxins, has remained unclear. In addition, since the VirR/VirS–VR-RNA cascade does not regulate all toxin genes, other factors should be involved in regulating the expression of these genes, although little is known about such regulators, except for VirR/VirS in C. perfringens.
The present study focuses on two putative transcriptional regulators with a DNA-binding motif that are expressed during the exponential growth phase. CPE1447 and CPE1446 have a helix-turn-helix (HTH) motif in the N-terminal amino acid sequence, suggesting that the proteins function as transcriptional factors. CPE1447 and CPE1446 regulate the expression of the toxin genes plc, pfoA, nanI, and nagHIJKat the transcriptional level. Since the regulation of these genes requires both CPE1447 and CPE1446, they cooperate to control the expression of the target genes. The present study also found that the CPE1447 and CPE1446 proteins form a stable complex in C. perfringenscells and that they regulate a different set of toxin genes than the VirR/VirS–VR-RNA regulatory cascade. These results demonstrate that CPE1447 and CPE1446 are new regulatory factors that are involved in toxin regulation in C. perfringensand that they are important for pathogenesis, along with the VirR/VirS–VR-RNA cascade.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. perfringensstrain 13 (15), the derivative strain TS140 (23), a CPE1446 mutant (NO13), and a CPE1447-CPE1446 deletion mutant (NO16) were cultured under anaerobic conditions using an Anaeropack (Mitsubishi Gas Chemical Co. Inc., Tokyo, Japan) at 37°C in Gifu anaerobic medium (GAM) broth or brain heart infusion (BHI)-sheep blood agar plates (Difco and Nippon Biotest Laboratories Inc.) with or without 50 μg/ml erythromycin. C. perfringensharboring plasmids was cultured in GAM broth containing 25 μg/ml chloramphenicol. Escherichia coliJM109 harboring plasmids was grown at 37°C in LB medium supplemented with either 50 μg/ml ampicillin or 25 μg/ml chloramphenicol.
Construction of CPE1446 mutant and CPE1447-CPE1446 deletion strains.
We inactivated the CPE1446 gene by a single-crossover event using a plasmid vector lacking the repgene of C. perfringensand constructed the CPE1446 disruptant NO13. An internal region of CPE1446 (497 bp) and the ermBPgene were amplified by PCR using primers NO1/NO2 and E1/E2 from the genomic DNA of C. perfringensstrain 13 and pJIR418 (24), respectively, and cloned into the PstI and EcoRI sites of pUC18, respectively. The resulting plasmid, pNO13, was introduced into C. perfringensstrain 13 by electroporation.
The upstream region of CPE1447 (921 bp) and the downstream region of CPE1446 (966 bp) were amplified by PCR using primers NO3/NO4 and NO5/NO6 from the genomic DNA of C. perfringensand digested with EcoRI-KpnI and PstI-BamHI, respectively. These DNA fragments were cloned into appropriately digested pUC18, and the resulting plasmid was designated pNO15. The ermBPgene, amplified from pJIR418 using primers E3/E4, was inserted into the BamHI site of pNO15, resulting in pNO16. To construct the CPE1447-CPE1446 deletion mutant strain NO16, over 6 μg of DNA fragments amplified from pNO16 using primers NO3/NO6 were introduced into C. perfringensstrain 13 by electroporation. We independently introduced this linear DNA fragment twice and obtained two transformants in each experiment. We used PCR and Southern blot analysis to confirm the correct insertion of the resistance gene and deletion of the CPE1447-CPE1446 gene in 3 of 4 transformants.
Electroporation proceeded as described below. C. perfringensstrain 13 was grown to the late-exponential phase in 10 ml of GAM broth. The cells were harvested by centrifugation, washed twice with 15 ml of 15% glycerol, and resuspended in 4 ml of 15% glycerol. The plasmid vectors or DNA fragments were mixed with 80 μl of cell suspension in a 2-mm-gap cuvette. Electroporation proceeded using a Bio-Rad Gene Pulser II (Bio-Rad Laboratories) set at a capacitance of 25 μF and a resistance of 200 Ω and using pulses of 2.5 kV (12.5 kV/cm). After pulsing, the cells were mixed immediately with 500 μl of GAM broth and incubated at 37°C for 2 h. The transformants were selected on BHI-sheep blood agar containing 50 μg/ml erythromycin. Each mutant was confirmed by PCR, Southern blot analysis, and DNA sequencing.
Plasmids.
We constructed the His-tagged CPE1446 and CPE1447 coexpression vector, pCPE6, as follows. A DNA fragment containing a CPE1447 promoter and the CPE1447 and CPE1446 open reading frame (ORF) was amplified by PCR using primers NO7/NO10 from the genomic DNA of C. perfringensstrain 13. The PCR product was digested with EcoRI and BamHI and then inserted into the same sites of the His-tagged Tex protein expression vector (K. Abe, personal communication).
The DNA fragments containing the CPE1447 promoter or CPE1446 ORF were amplified by PCR using primers NO7/NO8 and NO9/NO10, respectively. The PCR products were then digested with EcoRI or with EcoRI and BamHI and cloned into the same sites of the His-tagged Tex protein expression vector, resulting in pCPE9 expressing the CPE1446-His protein from the native promoter located upstream of the CPE1447 ORF.
The CPE1447-CPE1446 expression vector, pNOC16, derived from the pJIR418 vector, contained the DNA fragment amplified by PCR using primers NO11/NO6.
To construct the CPE1447–glutathione S-transferase (GST) expression vector pCPE43, a PCR fragment amplified using primers NO7/NO12 was inserted into the EcoRI-BamHI sites of pCPE33 (16).
The CPE1446-His or CPE1447 and CPE1446-His protein overexpression vectors, pNOE03 and pNOE05, respectively, were constructed as followed. Fragments amplified from the genomic DNA of C. perfringensstrain 13 by PCR using the NOE2/NO10 or NOE1/NO10 primers were cloned into the NcoI-BglII sites of pQE60.
Oligonucleotides.
Table S1 in the supplemental material lists the oligonucleotide primers used in this study.
Northern blot analysis.
Total RNA extracted from C. perfringensderivatives grown at 37°C in GAM broth were subjected to Northern blot analysis as described previously (16) with digoxigenin-labeled DNA probes generated by DIG-High Prime according to the supplier's instructions (19a). Template DNAs for the toxin gene-specific probes were amplified by PCR from strain 13 genomic DNA using the primers described by Abe et al. (1).
Primer extension analysis.
The 5′ ends of CPE1447, nagI, nagK, and nanImRNAs were determined using the PE1, PE2, PE3, and PE4 primers, respectively (see Table S1 in the supplemental material). The primers (10 pmol) were end labeled with 1 μl of T4 polynucleotide kinase (PNK), 2 μl of 10× PNK buffer, and 5 μl of [γ-32P]ATP in 20 μl of reaction buffer at 37°C for 30 min. The T4 PNK was then inactivated by heating it at 95°C for 2 min. The reaction mixture (10 μl) containing 8 μg of total RNA, 1 nmol of deoxynucleoside triphosphates (dNTPs), and 0.5 pmol of labeled primer was incubated at 95°C for 1 min, followed by 65°C for 5 min, and then cooled on ice. The extension reaction mixture was incubated at 42°C for 1 h with 20 units of RNase inhibitor (Takara Bio Ltd.) and 200 units of PrimeScript reverse transcriptase (Takara Bio Ltd.). cDNA products precipitated with ethanol were separated on 6% polyacrylamide–7 M urea–Tris-borate-EDTA (TBE) gels. Sequence ladders were generated using the fmol DNA sequencing system (Promega) according to the supplier's instructions.
Purification of His-tagged protein.
E. coliM15 harboring pREP4 (Qiagen) and pNOE03 or pNOE05 was grown in 100 ml of LB broth containing 50 μg/ml ampicillin and 25 μg/ml kanamycin to the mid-exponential phase. After adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), the cells were incubated for an additional 5 h and then harvested by centrifugation. The cells were then resuspended in 5 ml of lysis buffer containing 50 mM NaH2PO4, pH 8.0, 300 mM NaCl, and 10 mM imidazole and passed twice through a French press. After adding 300 μl of His-Select Nickel Affinity Gel (Sigma), the cell extract was gently mixed at 4°C for 1 h. The gel binding to the His-tagged protein was washed twice with lysis buffer containing 20 mM imidazole. The His-tagged protein was then eluted with lysis buffer containing 250 mM imidazole and dialyzed against dialysis buffer containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 50% glycerol, and 200 μM phenylmethylsulfonyl fluoride at 4°C for 16 h.
CPE1446-His protein was purified from C. perfringensas described above. NO16 harboring pCPE6 or pNOC16 (negative control for coprecipitation) was grown in GAM broth at 37°C to the mid-exponential phase. The cells were resuspended in 3 ml of lysis buffer and passed three times through a French press. The proteins were analyzed by 12% SDS-PAGE or by Western blotting using anti-His antibody.
Western blot analysis.
Whole proteins were extracted using glass beads. C. perfringensharboring the CPE1446-His or CPE1447-GST protein expression vector was cultured in 30 ml of GAM broth containing 25 μg/ml chloramphenicol at 37°C. Cells were harvested from 10 ml culture by centrifugation at 2, 3, and 4 h after inoculation. The cell pellets were resuspended in 1 ml of LETS buffer (100 mM LiCl, 10 mM EDTA, 10 mM Tris-HCl, pH 7.5, and 1% [wt/vol] sodium dodecyl sulfate), and then 200 μl of the suspension was mixed with 100 μl of glass beads and vortexed for 4 min. After centrifugation, the supernatants were collected. Equivalent volumes of 0.02 A280unit of protein samples were resolved by SDS-PAGE and then electroblotted onto polyvinylidene difluoride membranes. The membranes were blocked with 5% skim milk in Tris-buffered saline containing 0.2% Tween 80 and then probed with anti-His tag (Wako) diluted 1:5,000 or anti-DnaK diluted 1:5,000. Horseradish peroxidase-conjugated anti-mouse or -rabbit secondary antibodies (GE Healthcare) were applied at a dilution of 1:50,000. Bound antibodies were then detected using Immunostar LD (Wako).
RESULTS
CPE1446 and CPE1447 are cotranscribed.
CPE1446 and CPE1447 are located in a region downstream of the CPE1445 gene in the reverse direction (Fig. 1A). The CPE1447 locus was previously suggested to include the CPE1448 (528 bp) and CPE1447 (753 bp) genes in C. perfringensstrain 13 (22) (Fig. 1A). However, a BLAST search revealed that the CPE1448 and CPE1447 genes have sequence similarity to the N- and C-terminal regions, respectively, of an orthologous CPR_1434 gene in C. perfringensSM101, which is an enterotoxin-positive type A strain. In addition, the CPR_1434 gene is 1,302 bp long, which is almost equal to the combined lengths of CPE1448 and CPE1447. We constructed a His-tagged CPE1447 expression vector, which contained the whole CPE1449-CPE1448 intergenic region (IGR) (presumably containing the native promoter) and the CPE1448-CPE1447 locus. The histidine tag was fused to the C terminus of the CPE1447 ORF in the plasmid vector. If CPE1448 and CPE1447 encode their respective proteins, the CPE1447 protein should be roughly 29 kDa (250 amino acids [aa]). Western blot analysis using anti-His tag antibody revealed that the CPE1447-His protein was almost 50 kDa (data not shown). We then sequenced the region surrounding the CPE1448 stop codon and determined that the previously annotated TAA stop codon was a TCA codon encoding serine. This indicates that CPE1448 and CPE1447 encode a single protein comprising 434 amino acids. Here, we refer to the CPE1448-CPE1447 fusion gene as CPE1447 (Fig. 1A).
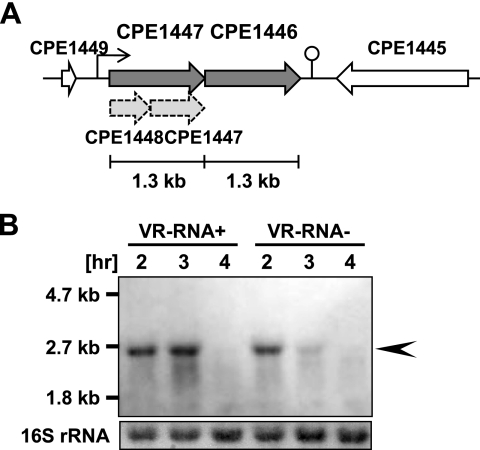
Fig. 1.
Expression of CPE1447-CPE1446 mRNA. (A) Schematic drawing of the CPE1447-CPE1446 locus in the C. perfringensgenome. The transcriptional start site and terminator are indicated by a bent arrow and a stem-loop structure, respectively. Previously annotated CPE1448 and CPE1447 genes are indicated by dashed arrows. (B) Northern blots of strains 13 (VR-RNA+) and TS140 (VR-RNA−) harboring the pJIR418 plasmid vector. Total RNA was isolated from cultures at the indicated time points, and 2 μg of total RNA was subjected to Northern blot analysis using a probe specific for the CPE1446 gene. The 16S rRNA on the blot detected by methylene blue staining is indicated at the bottom. The arrowhead indicates polycistronic mRNA encoding CPE1447 and CPE1446.
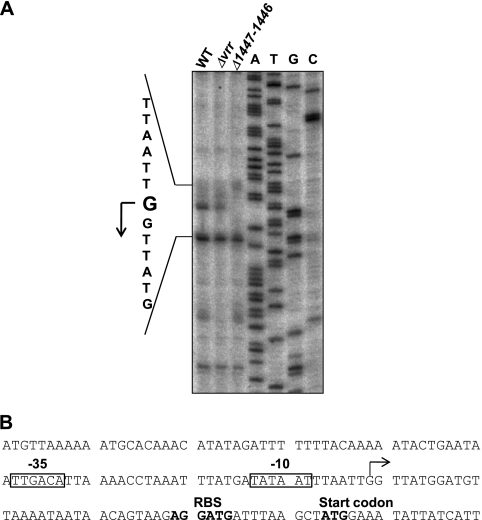
The CPE1446 and CPE1447 genes were separated by a 16-bp intergenic region, suggesting that the genes are cotranscribed as an operon. Northern blot analysis was carried out with total RNA from C. perfringensstrain 13 cells at various time points (2 to 4 h of culture). A 2.6-kb transcript was detected after 2 or 3 h in culture (Fig. 1B), suggesting that CPE1447 and CPE1446 were cotranscribed at the mid- and late-exponential phase. The CPE1447-CPE1446 operon is conserved in some Clostridiumspecies, such as C. acetobutylicum, C. botulinum, C. tetani, C. novyi, C. beijerinckii, and C. kluyveri(data not shown), suggesting that the operon plays an important role in gene regulation among clostridia. We performed primer extension analysis to determine the transcriptional start site of CPE1447-CPE1446 mRNA (Fig. 2A). Although several extension products were detected, only the transcriptional start site indicated in Fig. 2was detected by two independent primer extension analyses using different primers (data not shown). Thus, the extension products, except that of the transcriptional start site that we defined, might be nonspecific and caused by mispriming. A typical promoter sequence recognized by RNA polymerase σA(TTGACA[N]19TATAAT) was observed upstream of the proposed transcriptional initiation site (Fig. 2B).
Fig. 2.
Determination of the transcriptional start site located upstream of CPE1447. (A) Primer extension analysis proceeded using 32P-labeled primer and total RNA isolated from the cultures of strains 13, TS140, and NO16 at the mid-exponential phase. The sequence around the transcriptional start site for CPE1447 was determined by sequence reaction using the same primer and is indicated on the left. (B) DNA sequence around the CPE1447 promoter. The transcriptional start site of CPE1447 is indicated by a bent arrow. Putative −35 and −10 sequence regions are boxed. The ribosome binding site (RBS) and start codon of CPE1447 are shown in boldface.
CPE1446 and CPE1447 regulate the expression of several toxin genes in concert.
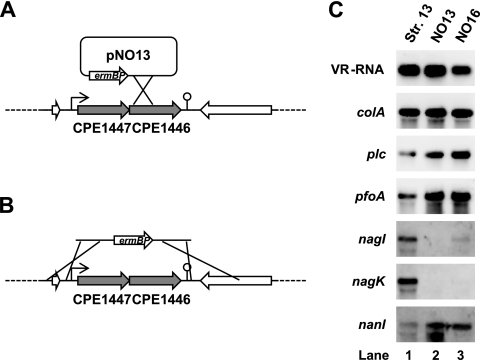
The CPE1446 and CPE1447 proteins possess the HTH motif in the N-terminal sequence and might function as DNA-binding proteins and transcriptional regulators. Northern blot analysis indicated that CPE1447-CPE1446 mRNA accumulated jn the mid- and late-logarithmic growth phase (Fig. 1B), when many genes, including those encoding toxins and extracellular enzymes, are expressed at high levels in C. perfringens(19). This suggests that CPE1447 and CPE1446 are involved in toxin gene regulation. To examine this notion, we constructed strain NO13, in which the CPE1446 gene was disrupted by a single-crossover event using a suicide vector (Fig. 3A), and strain NO16, a CPE1446 and CPE1447 deletion mutant (Fig. 3B). Both NO13 and NO16 were grown to the mid-logarithmic phase. Total RNAs isolated from the cultures were used for Northern blot analyses of the toxin genes. The colAgene, which encodes a collagenase, and VR-RNA, which regulates several toxin genes (16, 17, 23), are identically expressed in strains 13, NO13, and NO16 (Fig. 3C). VR-RNA directly regulates colAexpression through an antiantisense mechanism (16), and CPE1447 and CPE1446 were not associated with the expression of either gene. On the other hand, expression of the plc, pfoA, and nanIgenes, which encode phospholipase C, perfringolysin O, and sialidase, respectively, was activated in NO13 and NO16 compared to strain 13 (Fig. 3C). In addition, expression of the hyaluronidase genes nagIand nagKwas obviously repressed in NO13 and NO16 compared with strain 13 (Fig. 3C). These results suggest that CPE1447 and CPE1446 affect the expression of several toxin genes.
Fig. 3.
CPE1447 and CPE1446 regulate expression of several toxin genes. (A) Schematic drawing of suicide plasmid pNO13 integrated into the CPE1446 gene of C. perfringensstrain 13. The transcriptional start site and terminator are indicated by a bent arrow and a stem-loop structure, respectively. (B) Schematic drawing of a DNA fragment used to delete the CPE1447 and CPE1446 genes in C. perfringensstrain 13. The fragment contains upstream and downstream sequences of CPE1447 and CPE1446, respectively, and an erythromycin resistance gene. (C) Northern blots of strains 13, NO13, and NO16. Total RNA (1 μg) isolated from culture at the mid-exponential phase was resolved on 1% agarose gels containing 2% formaldehyde and then subjected to Northern blot analysis using toxin gene-specific probes.
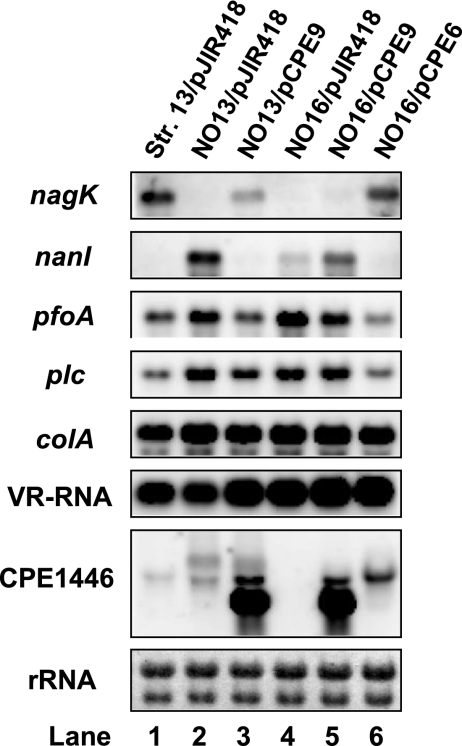
To further analyze the relationship between the CPE1447 and CPE1446 genes and toxin gene expression, CPE1447 and CPE1446 or CPE1446 expression vectors were constructed and introduced into NO13 and NO16. The activation and repression of the toxin genes in NO16 were restored by complementation with plasmid-encoded CPE1447 and CPE1446, but not with CPE1446 alone (Fig. 4; lanes 5 and 6). Transformation of the plasmid vector expressing CPE1446 into NO13 cells also restored toxin gene expression, suggesting that the CPE1446 gene carried in a plasmid is functionally equivalent to a chromosomally encoded copy (Fig. 4, lane 3). These results suggest that inactivation of either CPE1446 or CPE1447 causes changes in the expression of the toxin genes. Moreover, CPE1446 and CPE1447 would coordinately regulate several toxin genes.
Fig. 4.
CPE1447 and CPE1446 function in a coordinated manner. C. perfringensstrains 13, NO13, and NO16 harboring the CPE1446 or CPE1447 and CPE1446 expression vector were grown at 37°C to the mid-exponential phase. Total RNA (1 μg) isolated from the culture was used for Northern blot analysis with the indicated gene-specific probes. The 23S and 16S rRNAs on the blot detected by methylene blue staining are indicated at the bottom.
CPE1446 and CPE1447 proteins form a stable complex in vivo.
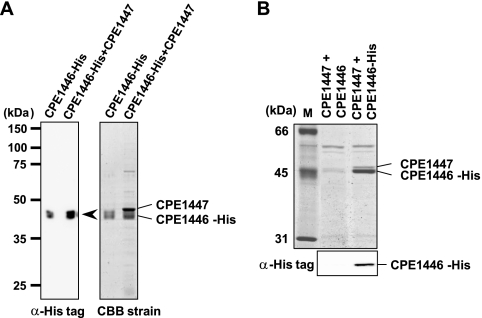
DNA-binding proteins with the HTH motif typically form an oligomer and bind to target DNA sequences. Neither the purified CPE1447 nor the CPE1446 protein alone could specifically bind to the target DNA sequences (nagI, nagK, and nanI) (data not shown), but they regulated common target genes in a coordinated manner. Therefore, we postulated that these proteins form a complex in vivo. To confirm interaction between CPE1447 and CPE1446, CPE1447 and C-terminally His-tagged CPE1446 proteins were coexpressed in E. coliusing a plasmid vector. Both proteins were simultaneously overexpressed, and the His-tagged CPE1446 protein was then purified and analyzed by SDS-PAGE (Fig. 5A). Purified CPE1446-His protein was detected using anti-His tag antiserum (Fig. 5A, left). When CPE1447 was concomitantly overexpressed, a second protein was copurified with CPE1446-His protein (Fig. 5A, right). This protein migrated slightly later than CPE1446-His protein and was not detected when only CPE1446-His was overexpressed. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis confirmed that the CPE1447 and CPE1446-His proteins coprecipitated (data not shown). To determine whether the CPE1446 and CPE1447 proteins also form a complex in their native host, C. perfringens, we introduced the CPE1447 and CPE1446-His protein expression vector, in which CPE1447–CPE1446-His mRNA transcription starts from the native promoter, into NO16, the CPE1447-CPE1446 operon deletion strain. The CPE1446-His and CPE1447 proteins from the vector functioned like C. perfringensgenomic CPE1446 and CPE1447, because the plasmid vector restored the toxin gene expression of NO16 (Fig. 4, lane 6). His-tagged protein was purified from cell extracts of C. perfringens, and the proteins were analyzed by SDS-PAGE (Fig. 5B). The purification of CPE1446-His protein was confirmed by Western blot analysis with anti-His tag antiserum (Fig. 5B, bottom). The CPE1447 protein was also copurified with CPE1446-His protein from cell extracts of C. perfringens, but not from cells expressing CPE1446 without a His tag (Fig. 5B, top). Therefore, the CPE1446-His and CPE1447 proteins interact and form a stable complex in their native host.
Fig. 5.
CPE1447 and CPE1446 form a stable complex in vivo. (A) Coprecipitation of coexpressed CPE1447 and CPE1446-His proteins in E. colicells. Coprecipitated His-tagged proteins (20 ng) were detected by Western blot analysis using anti-His tag antibody (left). Purified proteins (2 μg) were resolved by SDS-PAGE and stained with Coomassie brilliant blue (CBB) (right). (B) SDS-PAGE (top) and Western blots (bottom) of proteins coprecipitated with CPE1446-His protein from C. perfringenscells. The protein complexes were purified from extracts of C. perfringensNO16 cells harboring plasmids expressing CPE1447-CPE1446 or CPE1447 and His-tagged CPE1446. The SDS-PAGE gel was stained with Coomassie brilliant blue. The Western blots were probed with anti-His tag antibody.
The clostridial small regulatory RNA (sRNA), VR-RNA, is involved in CPE1446 and CPE1447 protein expression.
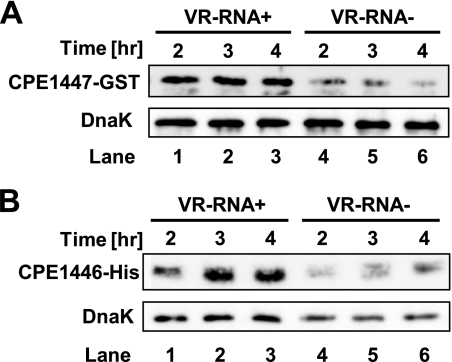
The VirR/VirS two-component system (TCS) is a global regulator in C. perfringens(3, 18). Because a VirR binding motif (CCAGTTNNNCAC) does not exist in the CPE1447 promoter sequence, the expression of CPE1447-CPE1446 is not directly regulated by the VirR response regulator (5, 6). Several genes, including those that encode toxins, are regulated by the VirR/VirS–VR-RNA cascade in C. perfringens(17). VR-RNA is a regulatory RNA that influences the expression of many genes, including several toxin genes, and might also regulate that of the CPE1447-CPE1446 genes. To determine whether CPE1447-CPE1446 expression is regulated by VR-RNA, we examined CPE1447-CPE1446 mRNA expression in TS140 cells, which are deficient in VR-RNA. Northern blots showed that CPE1447-CPE1446 mRNA levels decreased in TS140 cells and that VR-RNA positively regulates expression (Fig. 1B). This finding was consistent with the results of primer extension analysis, in which the amount of extension product was decreased in TS140 cells compared to strain 13 (Fig. 2A). To confirm that VR-RNA regulates CPE1447 and CPE1446, we constructed a CPE1447-GST translational fusion gene in which the CPE1447 promoter, the CPE1447 5′ untranslated region (UTR), and part of the CPE1447 ORF (region −45 to +45 relative to A of the AUG start codon) were fused in frame to the GST gene. A His tag was also fused to the C terminus of the CPE1446 codon of the CPE1447-CPE1446 operon in a plasmid vector in which CPE1447 and CPE1446-His proteins were expressed from the native promoter. The expression of both the CPE1447-GST and CPE1446-His proteins decreased in TS140 after 2 to 4 h in culture (Fig. 6). This indicated that VR-RNA activates the synthesis of both the CPE1447 and CPE1446 proteins in C. perfringens.
Fig. 6.
VR-RNA influences the amounts of CPE1447-GST and CPE1446-His proteins. (A) Western blots of strain 13 (VR-RNA+) or TS140 (VR-RNA−) harboring pCPE43. Each lane was loaded with 0.02 A280unit of protein obtained from cells at the indicated time points. A GST fusion protein and DnaK were probed with anti-GST and anti-DnaK antibody, respectively. (B) Western blots of strain 13 or TS140 harboring pCPE6. Each lane was loaded with 0.02 A280unit of protein from cells at the indicated time points. A His tag fusion protein and DnaK were probed with anti-His tag and anti-DnaK antibody, respectively.
CPE1447/CPE1446 and VR-RNA affect the expression of a different set of toxin genes.
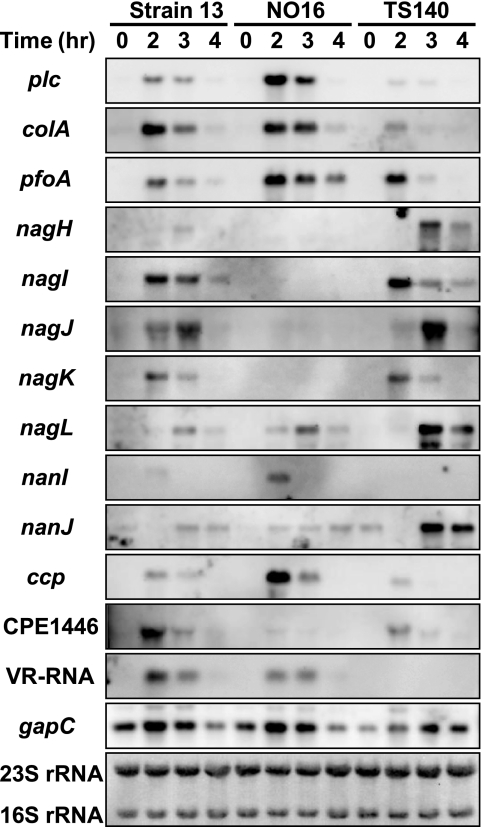
The present results suggested that VR-RNA and the CPE1447-CPE1446 protein complex form a regulatory cascade. In other words, VR-RNA affects the expression of the CPE1447-CPE1446 target genes by regulating the amount of CPE1447 and CPE1446 proteins. To prove this hypothesis, total RNA was isolated from cultures of C. perfringensstrains 13, NO16, and TS140 at various time points and used for Northern blot analysis of the toxin genes (Fig. 6). The plc, pfoA, nanI, and ccpmRNAs increased, whereas the nagH, nagI, nagJ, and nagKmRNAs decreased in NO16 compared to strain 13 (Fig. 7). C. perfringenspossesses five hyaluronidase and two sialidase genes (22), four and one of which, respectively, are regulated by CPE1447-CPE1446. On the other hand, plcand colAmRNAs were repressed while nagH, nagL, and nanJmRNAs were activated in TS140, as previously reported (17). If VR-RNA regulates the toxin genes by controlling CPE1447 and CPE1446 protein production, CPE1447-CPE1446 target toxin genes should be similarly expressed in both NO16 and TS140. However, CPE1447-CPE1446 positively and negatively regulated, whereas VR-RNA repressed and activated, respectively. Thus, VR-RNA and CPE1447-CPE1446 do not influence the expression of CPE1447-CPE1446 target genes in the same manner and would influence the expression of different sets of toxin genes.
Fig. 7.
CPE1447-CPE1446 and VR-RNA independently regulate the target genes. Northern blot analysis of strains 13, NO16, and TS140 was performed with the indicated gene-specific probes. C. perfringenswas grown and harvested at the indicated time points. Total RNA (1 μg) isolated from cultures was subjected to Northern blot analysis. The 16S and 23S rRNAs on the blot detected by methylene blue staining are indicated at the bottom.
Determination of the transcriptional start sites of CPE1447-CPE1446 target genes.
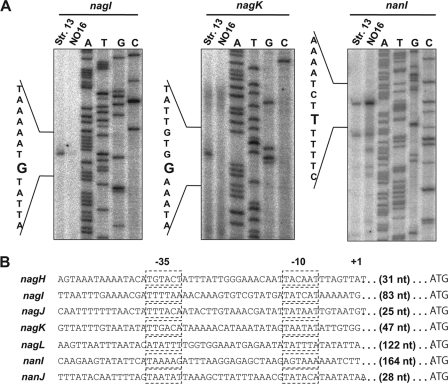
We investigated the transcriptional start sites of the CPE1447-CPE1446 target genes, nagI, nagK, and nanI. Total RNAs isolated from strains 13 and NO16 at the mid-exponential phase were used for primer extension analyses (Fig. 8A). The nagI, nagK, and nanItranscriptional start sites were located 83, 47, and 164 nucleotides (nt) upstream of the AUG start codon, respectively. Several extension products of the nanItranscripts were observed, suggesting that nanImRNA is transcribed from multiple promoters. However, because the longer transcripts of nanImRNA shown in Fig. 8A accumulated to high levels and increased in the CPE1447-CPE1446 deletion strain, we defined the 5′ end of this extension product as the major CPE1447-CPE1446-dependent transcriptional start site. The canonical recognition sequences of RNA polymerase containing σAwere TATAAT for the “−10 box” and TTGACA for the “−35 box.” These consensus sequences were identified upstream of the nagIand nagKtranscriptional start sites (Fig. 8B). Although, the high AT content of the C. perfringensgenomic sequence sometimes results in poor similarity between the −10 and −35 regions of the promoter and the consensus sequences (12), the nanIpromoter sequence is less effectively recognized by σARNA polymerase holoenzyme, suggesting that nanItranscription depends on an alternative sigma factor (Fig. 8B). The cDNA extension products of nagIand nagKin NO16 decreased, and that of nanIincreased. These finding were consistent with those of the Northern blots (Fig. 8A). A comparison of the promoter sequences of the CPE1447-CPE1446 target genes revealed no sequence similarity.
Fig. 8.
Determination of transcriptional start sites of nagI, nagK, and nanI. (A) Primer extension analysis of nagI, nagK, and nanIusing 32P-labeled primer and total RNA isolated from cultures of strains 13 and NO16 at the mid-exponential phase. The sequence around the transcriptional start site of each gene was determined by sequence reaction using the same primer. The transcriptional start sites are shown on the left. (B) Promoter sequences of hyaluronidase and sialidase genes in C. perfringensstrain 13 (1). The dashed boxes indicate −10 and −35 boxes. The ranscriptional start site is indicated by +1′. The length of the 5′ UTR of each gene is also indicated.
DISCUSSION
The present study showed that CPE1447 and CPE1446 with an HTH domain at the N terminus are novel regulators of toxin gene expression in C. perfringens. Inactivation of the CPE1446 gene and deletion of the CPE1447-CPE1446 operon influenced expression of the same set of toxin genes (Fig. 3). Furthermore, coexpression of CPE1447 and CPE1446 is required to regulate these genes, suggesting that CPE1447 and CPE1446 coordinately function as toxin regulators (Fig. 4). Coprecipitation assays of His-tagged CPE1446 protein revealed interactions between the CPE1447 and CPE1446 proteins in vivo, indicating that CPE1447 and CPE1446 form a stable heteromeric complex (Fig. 5). Therefore, we considered that a heteromeric oligomer of CPE1447 and CPE1446 binds to the target promoter and regulates transcription in C. perfringens.
The CPE1447 and CPE1446 amino acid sequences downstream of the HTH motif contain several tetratricopeptide repeat (TPR) domains, which form a coiled-coil or helical-bundle structure (9). Protein-protein interactions such as those between DnaJ-like and Hsp70 proteins are mediated by TPR motifs (8). Thus, TPR motifs in the CPE1447 and CPE1446 proteins would enable them to form heteroprotein complexes. CPE1447 or CPE1446 purified from E. coliitself did not specifically bind to the promoter sequences of nagI, nagK, and nanIin gel mobility shift assays (electrophoretic mobility shift assays [EMSA]) (data not shown), implying that heteromeric complex formation is critical for binding to the target sequence of CPE1447 or CPE1446. We analyzed interactions between the DNA sequences of the nagI, nagK, and nanIpromoters and the CPE1447-CPE1446 protein complexes using an EMSA. The protein complexes were purified by coprecipitating CPE1447 and CPE1446-His proteins from E. coli(Fig. 5A). Upstream regions of transcriptional start sites about 100 bp long were amplified by PCR and end labeled with [α-32P]ATP. When 15 fmol of DNA probe was mixed with 1 μg of purified CPE1447-CPE1446 protein complex for 30 min at 37°C, we could not observe an obvious shift band representing DNA-protein complexes, although the signal of the free DNA probe slightly decreased. The VirR protein is a DNA-binding transcriptional regulator of C. perfringens, and 1 μg of the protein purified from E. coliforms DNA-protein complexes with its target pfoApromoter in vitro(5, 6). These results suggest that the CPE1447-CPE1446 protein complex purified from E. colidoes not bind the nagI, nagK, and nanIpromoter with high affinity. We also analyzed cell lysates from C. perfringensusing EMSA with DNA fragments of the whole 5′ IGR of the nagI, nagK, and nanIgenes. Several DNA-protein complexes were detected, suggesting that some proteins in the cell lysate interact with these DNA sequences. However, disruption of the CPE1446 gene did not alter the shift band in EMSA, indicating that the protein(s) that bound to the DNA in the EMSA was not CPE1446. Therefore, direct evidence of CPE1447 and CPE1446 binding to DNA fragments, including the target promoter and its upstream region, was not obtained. In addition, a comparison of the promoter sequences of nagHIJKand nanIdid not find similar sequences (Fig. 8). These toxin genes might not be the direct targets, or CPE1447 and CPE1446 might not function as DNA-binding proteins, although both CPE1447 and CPE1446 have an HTH domain. However, we consider that another factor is involved in regulation by the CPE1447-CPE1446 protein complex in C. perfringenscells.
This study found that the expression of eight genes encoding toxins was influenced in a CPE1447 and CPE1446 deletion strain, suggesting that the CPE1447-CPE1446 protein complex is a global regulator in C. perfringens. The CPE1447 and CPE1446 genes are widely conserved in clostridia, including C. acetobutylicum, C. beijerinckii, C. botulinum, C. tetani, C. novyi, C. kluyveri, C. ljungdahlii, and C. cellulovorans. Among these, C. botulinum, C. tetani, and C. novyiare causative agents of botulism, tetanus, and gas gangrene, respectively, in humans. On the other hand, C. acetobutylicum, C. beijerinckii, C. kluyveri, C. ljungdahlii, and C. cellulovoransproduce useful solvents or enzymes and should have industrial value. Thus, both pathogenic and industrially interesting clostridial species possess this heteromeric protein complex regulator. Identification of a precise CPE1447-CPE1446 regulon would also contribute to understanding the regulation of virulence factors and of useful genes in these clostridia that synthesize solvents and enzymes.
The clostridial sRNA, VR-RNA, which is a global regulator under the VirR/VirS two-component system, upregulates the expression of CPE1447-CPE1446 (Fig. 1and 6). However, Northern blot analysis of the CPE1447-CPE1446-null mutant and a VR-RNA-deficient mutant indicated that CPE1447-CPE1446 and VR-RNA affect the expression of a different set of the toxin genes (Fig. 7). We consider why VR-RNA does not affect CPE1447-CPE1446 target gene expression like CPE1447-CPE1446 while it positively regulates CPE1447-CPE1446 expression. One possibility is that a small amount of CPE1447-CPE1446 protein complex is sufficient to regulate the transcription of the target toxin genes. The CPE1447-CPE1446 deletion mutant and the CPE1446 disruptant have no CPE1447-CPE1446 protein complex. In contrast, CPE1447-CPE1446 expression is not completely repressed in the VR-RNA deletion mutant, in which low levels of CPE1447-CPE1446 protein complexes regulate target gene expression. A high level of CPE1447-CPE1446 expression from the plasmid vector complemented the toxin gene expression in a CPE1447-CPE1446 mutant strain (Fig. 4, lane 6) but did not excessively regulate these genes (that is, expression of the toxin genes did not differ between the wild type and a strain harboring the CPE1447-CPE1446 expression vector). This finding implies that the amount of CPE1447-CPE1446 proteins might not be involved in the regulatory efficiency of the target gene. Although the expression of over 100 genes changes in TS140 (17), a direct target gene of VR-RNA other than the collagenase gene, colA, is unknown (16). This suggests that the repression of CPE1447 and CPE1446 in TS140 is an indirect effect of the VR-RNA deletion. The expression of CPE1447 and CPE1446 transiently increases from the mid- to late-exponential phase (Fig. 1) and might be triggered by external signals. Thus, we propose that the VirR/VirS–VR-RNA cascade and CPE1447-CPE1446, respectively, regulate different subsets of genes in response to different signals in C. perfringens.
To date, only a few transcriptional regulators have been identified in C. perfringens(4, 13, 25). The present study discovered that the CPE1447-CPE1446 complex conserved across many clostridial species is a novel toxin gene regulator, and we also identified a possible new regulatory cascade. Further functional analysis of CPE1447 and CPE1446 is necessary to identify the direct target genes, as well as additional factors involved in the DNA-binding activity of CPE1447-1446 and external signals activating CPE1447-CPE1446 expression. Although further functional analysis of CPE1447 and CPE1446 is necessary, CPE1447-CPE1446 might be one of the most important regulatory factors among the pathogenic or industrially important Clostridiumspecies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nobuhiko Nomura for helpful discussions. We also thank Norma Foster for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Abe K., Obana N., Nakamura K. 2010. Effects of depletion of RNA-binding protein Tex on the expression of toxin genes in Clostridium perfringens. Biosci. Biotechnol. Biochem. 74:1564–1571 [DOI] [PubMed] [Google Scholar]
- 2. Awad M. M., Bryant A. E., Stevens D. L., Rood J. I. 1995. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15:191–202 [DOI] [PubMed] [Google Scholar]
- 3. Banu S., et al. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 35:854–864 [DOI] [PubMed] [Google Scholar]
- 4. Ba-Thein W., et al. 1996. The virR/virSlocus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 178:2514–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheung J. K., Dupuy B. D., Deveson S., Rood J. I. 2004. The spatial organization of the VirR boxes is critical for VirR-mediated expression of the perfringolysin O gene, pfoA, from Clostridium perfringens. J. Bacteriol. 186:3321–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheung J. K., Rood J. I. 2000. The VirR response regulator from Clostridium perfringensbinds independently to two imperfect direct repeats located upstream of the pfoApromoter. J. Bacteriol. 182:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiarezza M., et al. 2009. The NanI and NanJ sialidases of Clostridium perfringensare not essential for virulence. Infect. Immun. 77:4421–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cyr D. M., Langer T., Douglas M. G. 1994. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19:176–181 [DOI] [PubMed] [Google Scholar]
- 9. D'Andrea L. D., Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655–662 [DOI] [PubMed] [Google Scholar]
- 10. Davies J., et al. 1999. Reduction in the adherence of Pseudomonas aeruginosato native cystic fibrosis epithelium with anti-asialoGM1 antibody and neuraminidase inhibition. Eur. Respir. J. 13:565–570 [DOI] [PubMed] [Google Scholar]
- 11. Galen J. E., et al. 1992. Role of Vibrio choleraeneuraminidase in the function of cholera toxin. Infect. Immun. 60:406–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graves M. C., Rabinowitz J. C. 1986. In vivo and in vitro transcription of the Clostridium pasteurianumferredoxin gene. Evidence for “extended” promoter elements in gram-positive organisms. J. Biol. Chem. 261:11409–11415 [PubMed] [Google Scholar]
- 13. Hiscox T. J., et al. 2011. Regulation of virulence by the RevR response regulator in Clostridium perfringens. Infect. Immun. 79:2145–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hynes W. L., Walton S. L. 2000. Hyaluronidases of Gram-positive bacteria. FEMS Microbiol. Lett. 183:201–207 [DOI] [PubMed] [Google Scholar]
- 15. Mahony D. E., Moore T. I. 1976. Stable L-forms of Clostridium perfringensand their growth on glass surfaces. Can. J. Microbiol. 22:953–959 [DOI] [PubMed] [Google Scholar]
- 16. Obana N., Shirahama Y., Abe K., Nakamura K. 2010. Stabilization of Clostridium perfringenscollagenase mRNA by VR-RNA-dependent cleavage in 5′ leader sequence. Mol. Microbiol. 77:1416–1428 [DOI] [PubMed] [Google Scholar]
- 17. Ohtani K., et al. 2010. Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 16:258–264 [DOI] [PubMed] [Google Scholar]
- 18. Okumura K., Ohtani K., Hayashi H., Shimizu T. 2008. Characterization of genes regulated directly by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 190:7719–7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petit L., Gibert M., Popoff M. R. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104–110 [DOI] [PubMed] [Google Scholar]
- 19a. Roche Diagnostics, Inc 2000. DIG application manual for filter hybridization. Roche Diagnostics, Mannheim, Germany [Google Scholar]
- 20. Rood J. I., Cole S. T. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55:621–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rood J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333–360 [DOI] [PubMed] [Google Scholar]
- 22. Shimizu T., et al. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shimizu T., Yaguchi H., Ohtani K., Banu S., Hayashi H. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 43:257–265 [DOI] [PubMed] [Google Scholar]
- 24. Sloan J., et al. 1992. Construction of a sequenced Clostridium perfringens-Escherichia colishuttle plasmid. Plasmid 27:207–219 [DOI] [PubMed] [Google Scholar]
- 25. Varga J., Stirewalt V. L., Melville S. B. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 186:5221–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.