Abstract
We report expression and mutant phenotypes for a gene cluster in Sinorhizobium meliloti, designated cbtJKL, that has been shown to encode an ABC-type cobalt transport system. Transcription of cbtJKLinitiated 384 nucleotides upstream from the cbtJtranslation start codon, and the resulting 5′ region contained a putative B12riboswitch. Expression of the cbtJKLgenes appeared to be controlled by (cobalt-loaded) cobalamin interacting at the B12riboswitch, since (i) a putative B12riboswitch was located within this large upstream region, (ii) cbtJtranscription was repressed upon addition of cobalt or vitamin B12, and (iii) deletions in the B12riboswitch resulted in constitutive cbtJKLtranscription. Insertion mutants in cbtJKLfailed to grow in LB medium, and growth was restored through the addition of cobalt but not other metals. This growth phenotype appeared to be due to the chelation of cobalt present in LB, and cbtJKLmutants also failed to grow in minimal medium containing the chelating agent EDTA unless the medium was supplemented with additional or excess cobalt. In uptake experiments, 57Co2+accumulation was high in wild-type cells expressing the cbtJKLgenes, whereas wild-type cells in which cbtJKLexpression was repressed showed reduced accumulation. In cbtJKLmutant cells, 57Co2+accumulation was reduced relative to that of the wild type, and presumably, this residual cobalt transport occurred via an alternate ion uptake system(s) that is not specific to cobalt. In symbiosis, the alternate system(s) appeared to mediate cobalt transport into bacteroid cells, as low cbtJKLexpression was detected in bacteroids and cbtJKLmutants formed N2-fixing nodules on alfalfa.
INTRODUCTION
Cobalt is an essential trace element for many living organisms, as it plays a key biological role as the centrally coordinated ion in cyclic tetrapyrroles known as corrin rings (19, 40). Corrinoids, including the coenzyme vitamin B12(adenosylcobalamin [AdoCbl]) and its cobalamin (Cbl) derivatives, are coenzymes in a number of central metabolic reactions. Cobalt can also be associated directly with cobalt-dependent enzymes (noncorrin enzymes) (32). To acquire sufficient cobalt for metabolism, bacteria have high-affinity uptake systems to scavenge Co2+from the environment, where it is often available only in trace amounts (16, 56, 57, 69). When external metal concentrations are very high, Co2+accumulation may become toxic, and excess Co2+can be removed from cells by efflux systems (43, 63).
The NikMNQO and CbiMNQO uptake systems preferentially transport Ni and Co, respectively, and are common among bacteria and archaea (see references 55to 58and 74). These systems are members of a class of modular transporters which have substrate-specific components that are integral membrane proteins (CbiMN), energy coupling factor (ECF) transporters that consist of an ATPase typical of the ATP binding cassette (ABC) superfamily (CbiO), and a characteristic transmembrane protein (CbiQ) (57). These systems lack a periplasmic/extracellular substrate binding protein (11).
There are also a number of secondary, non-ABC-type Ni2+/Co2+transporters (16) whose metal ligand preference correlates in many cases with the genomic localization of the transport genes, whether they are adjacent to clusters for Ni- or Co-containing enzymes or to those for enzymes involved in Cbl biosynthesis (27, 55). For example, the Rhodococcus rhodochrous nhlFgene, encoding cobalt permease, lies beside the gene encoding a nitrile hydratase that contains a noncorrin Co2+(13, 33). Other divalent cation transport systems, such as ZupT (25) and Mg2+transport systems (CorA) (64), can also mediate Co2+uptake, but these are not likely to be physiologically relevant because of their poor affinity for Co2+.
Various bacteria are known to take up cobalamins from the environment, and the system of Salmonella entericaserovar Typhimurium and Escherichia coliconsists of an outer membrane TonB-dependent transporter, BtuB (2), the ABC-type transport proteins BtuC and BtuD, and the periplasmic binding protein BtuF (5). Genetically, btuFis not linked to the btuCEDoperon (6). BtuC is the transmembrane component and BtuD the ATP-binding component. The function of BtuE is uncertain and not required for vitamin B12transport (54). Although the btuFand btuCDgenes are often annotated in bacteria, sequence similarities among ABC-type siderophore/heme/vitamin B12family transport systems (34) make it difficult to identify the substrate(s) transported by these systems (74).
Riboswitches are conserved RNA elements in the 5′-untranslated region (5′-UTR) of prokaryotic mRNA molecules that modulate transcription attenuation or translation attenuation through the binding of specific effectors, such as vitamin B12, lysine, glycine, adenine, guanine, or glucosamine-6-phosphate (71). The regulated genes are usually involved in the biosynthesis or transport of the particular effector metabolite. For example, riboswitch sequences are often found 5′ of the cbiMNQOgenes (74). In E. coliand S. enterica, a B12riboswitch represses both transcription and translation of btuB(23, 50, 51), and conserved elements of the B12riboswitch are found 5′ of the cobalamin biosynthesis (cob) operon (42). Within the B12riboswitch region, there is a conserved motif called the B12box that is essential for AdoCbl-dependent regulation (44, 45, 52).
Sinorhizobium melilotiis a Gram-negative alphaproteobacterium that forms N2-fixing root nodules on its plant host, alfalfa. Co2+is required for the growth of S. melilotiand other rhizobia (36, 70) and is required for efficient nitrogen fixation in the Sinorhizobium-alfalfa symbiosis (15). S. melilotisynthesizes vitamin B12, and the Cbl-dependent enzymes methylmalonyl-coenzyme A (CoA) mutase, methionine synthase, and ribonucleotide reductase have been identified (9, 10, 14, 30, 62), as well as putative Cbl biosynthetic genes (4). The S. melilotiBluB protein was recently shown to catalyze a missing step in vitamin B12synthesis by cannibalizing flavin to form 5,6-dimethylbenzimidazole, the lower ligand of vitamin B12(7, 67). Recently, Co2+was observed to be required for survival of an S. melilotimutant that lacks the phosphotransferase system enzyme Hpr (48). BioM (CbiO homolog) and BioN (CbiQ homolog) in S. melilotihave been reported to import biotin (18), and these along with BioY appear to represent an ECF-type biotin transporter. A putative ABC-type cobalamin transporter (BtuF [Smb20056], BtuC [Smb20057], and BtuD [Smb20058]) was identified on the basis of sequence similarity to the siderophore/heme/vitamin B12family and the presence of a B12riboswitch in the 5′-UTR (69). Here we report that the ABC-type system encoded by the smb20056, smb20057, and smb20058genes transports Co2+but not cobalamin. We demonstrate that this transport system is required for growth of free-living cells at trace element concentrations of cobalt and that it is not required for symbiotic N2fixation in S. meliloti. We designate the transport system genes cbtJ(smb20056), cbtK(smb20057), and cbtL(smb20058), and we present expression data suggesting that cbtJKLexpression is repressed by Co2+and cobalamin via a 5′ B12riboswitch.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. E. colicultures were grown at 37°C in Luria broth (LB), and S. meliloticells were grown at 30°C in LB containing 2.5 mM CaCl2and 2.5 mM MgSO4. The defined M9-succinate medium contained 1× M9 salts (Difco) supplemented with 0.25 mM CaCl2, 1 mM MgSO4, 0.5 μg ml−1biotin, and 43 nM CoCl2(10 ng CoCl2·6H2O/ml) (60), with 15 mM succinate as the sole carbon source. MOPS (morpholinepropanesulfonic acid)-buffered minimal medium was used as previously described (73). One milliliter of a 1,000× trace element solution was added per liter of minimal medium. The trace element solution (1,000×) consisted of the following amounts of compounds per liter of H2O: 1.0 g H3BO3, 1.0 g ZnSO4·7H2O, 0.5 g CuSO4·5H2O, 0.5 g MnCl2·4H2O, 1.0 g NaMoO4·2H2O, 10.0 g EDTA, and 2.0 g NaFe-EDTA. We note that the 43 nM CoCl2that is routinely added to our M9 and MOPS minimal media (73) is in excess of that required for growth of cbtJKLmutants, and this accounts for the growth of cbtJKLmutant transconjugants on M9-succinate medium that is shown in Fig. 1A. Inductively coupled plasma-mass spectrometry (ICP-MS) analysis of the minimal medium employed in our experiments, with no added CoCl2, detected 2 nM cobalt.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic or genotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. colistrains | ||
| DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1Δ(argF-lacZYA) | Lab collection |
| MT616 | Conjugation helper strain carrying pRK600 | 20 |
| S. melilotistrains | ||
| RmP110 | Rm1021 with changed wild-type pstC; Smr | 74 |
| RmP831 | RmP110::pTH1968 cbtJp::gfp+-lacZSmrGmr(CbtJ+CbtK+CbtL+) | This study |
| RmP833 | RmP110::pTH1969 cbtJ::gfp+-lacZSmrGmr(CbtJ−CbtK−CbtL−) | This study |
| RmP835 | RmP110::pTH1970; 3′ end of cbtK::gfp+-lacZ; SmrGmr(CbtJ+CbtK+CbtL+) | This study |
| RmP889 | RmP110::pTH2030 cbtL::gfp+-lacZSmrGmr(CbtJ+CbtK+CbtL−) | This study |
| RmFL3108 | RmP110::pFL3108 cbtK::gfp+-lacZSmrGmr(CbtJ+CbtK−CbtL−) | This study |
| RmP1477 | RmP110::cobT::pTH2293 SmrGmr | This study |
| RmP2361 | RmP110 ΔB152 (pSymB Δ61,240–74,302), i.e., ΔcbtJKL | B. Peduska and T. M. Finan |
| RmP2364 | RmP2361(pTH2653) | Peduska and Finan |
| Plasmids | ||
| pFL3108 | pTH1522 carrying 3′ end of cbtK::gfp+-lacZand 5′ end of cobB; Gmr | 8 |
| pFL3283 | pTH1522 carrying cobT::gfp+-lacZ; Gmr | 8 |
| pTH1703 | Transcriptional reporter plasmid; Gmr | 8 |
| pTH1919 | pBBRMCS-3 with tetR-tetA(NsiI-BglII) from RK2 | This study |
| pTH1968 | pTH1703 carrying cbtJp(P20056F-P20056R); Gmr | This study |
| pTH1969 | pTH1703 carrying cbtJ::gfp+-lacZ(20056intF-20056intR); Gmr | This study |
| pTH1970 | pTH1703 carrying 3′ end of cbtL::gfp+-lacZ(20058endF-20058endR); Gmr | This study |
| pTH2030 | pTH1703 carrying cbtK::gfp+-lacZ(20058intF-20058intR); Gmr | This study |
| pTH2213 | pTH1919 derivative with ΔBglII; Tcr | This study |
| pTH2221 | pTH2213 carrying Ω terminator (ML18596-ML18597); Tcr | This study |
| pTH2224 | pTH2221 carrying gfp+-lacZ; Tcr | This study |
| pTH2237 | pTH2224 carrying cbtJp::gfp+-lacZ(ML18589-ML18594); Tcr | This study |
| pTH2238 | pTH2224 carrying cbtJp-5′-UTR 5′-UTR::gfp+-lacZ(ML18595-ML18589) | This study |
| pTH2239 | pTH2224 carrying cbtJp::gfp+-lacZ(MLML18590-ML18594); Tcr | This study |
| pTH2240 | pTH2224 carrying 5′-UTR and 5′ region of cbtJ(ML18590-ML18594); Tcr | This study |
| pTH2256 | pTH2224 carrying cbtJp-B12 riboswitch::gfp+-lacZ(ML18561-ML18589, ML18560-ML18594, and ML18589-ML18594) | This study |
| pTH2270 | pTH1703 carrying smb20060::gfp+-lacZ(B20059F-ML19261); Gmr | This study |
| pTH2293 | pFL3283 with Δgfp+-lacZ; Gmr | This study |
| pTH2303 | pTH2224 carrying cobPp(ML20074-ML20075); Tcr | This study |
| pTH2653 | pLAFR1 carrying positions 56,613 to 81,543 of pSymB (i.e., includes the cbtJKLgenes); Tcr | Peduska and Finan |
For plasmids, the primer pairs used to amplify genes are indicated in parentheses. Gm, gentamicin; Sm, streptomycin; Tc, tetracycline; Cm, chloramphenicol.
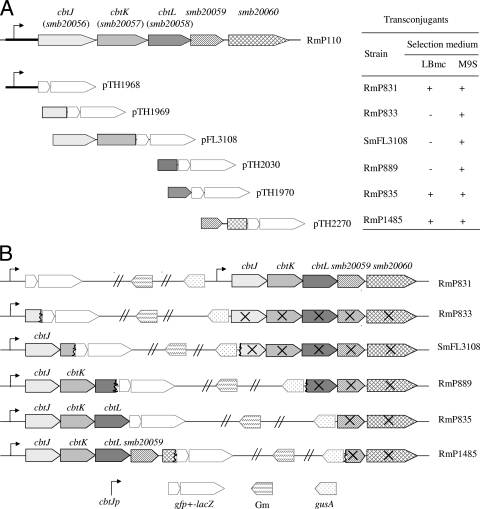
Fig. 1.
Schematic of the cbtJ-smb20060region on the pSymB megaplasmid of S. meliloti. (A) DNA fragments were PCR amplified and cloned upstream of the promoterless gfp+-lacZgenes in pTH1703 to obtain pTH1968, pTH1969, pTH2030, pTH1970, and pTH2270. Plasmid pFL3108 was a pTH1522 derivative carrying a genomic DNA fragment (8). Upon integration of the reporter fusion clones into the S. melilotigenome, transconjugant colonies grew (+) or did not grow (−) when selected on LB or M9-succinate containing streptomycin and gentamicin (Gm). Strains RmP831, RmP833, SmFL3108, RmP889, RmP835, and RmP1485 are representative transconjugant colonies selected for each integrated plasmid. (B) Diagram showing the genome organization of strains RmP831, RmP833, SmFL3108, RmP889, RmP835, and RmP1485 following recombination of the reporter plasmids. X's indicate the genes whose transcription was disrupted in each strain.
Antibiotics were used at the following concentrations: streptomycin, 200 μg ml−1; gentamicin, 60 μg ml−1(10 μg ml−1for E. coli); and tetracycline, 10 μg ml−1(15 μg ml−1for E. coli).
To identify substances required for growth of S. melilotimutants in LB, cells were initially grown for 36 h in M9-succinate medium, washed three times with 0.85% NaCl, and then inoculated into LB (optical density at 600 nm [OD600], ∼0.01). Following incubation for 16 h, the LB-grown cells were used to inoculate fresh LB (OD600, ∼0.01) supplemented alternatively with each of the ingredients used in M9 medium. The subcultures were grown for 16 h, and growth rates were monitored by measuring the OD600. To investigate whether Co2+was required for growth in minimal medium, S. melilotistrains were grown in LB with 5 μM CoCl2, washed three times with 0.85% NaCl, and then subcultured in MOPS medium (1) containing 2 mM inorganic phosphate (MOPS-P2) with CoCl2(0 to 20 nM). Subsequently, these MOPS-P2 cultures were subcultured again into MOPS-P2 medium with 0 to 20 nM CoCl2, and growth (OD600) was monitored.
DNA and RNA manipulations and microarrays.
DNA isolation, transformation, restriction, and ligation were performed by standard procedures (61). Oligonucleotide synthesis (Table 2) and DNA sequencing were performed at MobixLab (McMaster University, Hamilton, Ontario, Canada). To identify the cbtJtranscriptional start site through primer extension reactions, total RNA was isolated from RmP110 grown in LB or M9-succinate medium as described previously (73). Primers ML18489 and ML18490 were end labeled with [γ-32P]ATP, and following the primer extension reaction, the product was loaded onto a 6% acrylamide–7 M urea sequencing gel and electrophoresed alongside a sequencing ladder generated by using the same primer with plasmid pTH2240 as the DNA template.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a |
|---|---|
| P20056F | ATCTCTCGAGGACGAACGTAATAGTATAAC |
| P20056R | TCATAGATCTTCCCCGAGTGTGAGGCCGCT |
| 20056intF | GACACTCGAGAATTGCGGACGGCAGATCAC |
| 20056intR | GGCCAGATCTGCCTCTTCTCGGCTTTTCAG |
| 20058intF | GACTCTCGAGGCGAGCGGGGTTTCATGGTC |
| 20058intR | GATCAGATCTCCGATGTCGAGATGGTTGGT |
| 20058endF | TCGCCTCGAGCGCCGGAGGACGATGCGATC |
| 20058endR | GTCCAGATCTCAGGCTCCGACGGCGGCGATTG |
| B20059F | GGAAGTCGACGGTCCGTTTCATACTGGCCG |
| ML19261 | TTCAAGATCTCGTCCAGCCGCTTGTTGATC |
| ML18489 | ATGAGCGGTCGTTTCAGAAAGCTGTTCATG |
| ML18490 | ACAGTTGCGGGGGCAGCCACGGTTTTGGTC |
| ML18589 | GAGCACTAGTACAGGCATTCCCCCATACAT |
| ML18590 | CCGAACTAGTCTGTTGACGAACGTAATAGT |
| ML18593 | CTTCGGATCCGCTAGCCTTGGCAAGTACGCTTTCGAAGCT |
| ML18594 | GTTTGGATCCGCTAGCCATGAATAGTCCCCGAGTGT |
| ML18595 | GGGTGGATCCGCTAGCGGTCCCAGTTAATGTGGAAC |
| ML18596 | TAACGAGCTCCGGTGGATGACCTTTTGAAT |
| ML18597 | CCGGTCTAGAGGTGATTGATTGAGCAAGCT |
| ML19560 | CCGTCACGACGTTGGATCGATTTGGTCCCAGTTAATGTGGAACGC |
| ML19561 | GTTCATTAACTGGGACCAAATCGATCCAACGTCACGGGC |
| ML20074 | GCGCGAATTCCTGATCCGGACCGCTT |
| ML20075 | CGAGGGTACCGCATGCCATTACCGCTTGCCATAGCG |
Restriction sites that were engineered into the primers are underlined.
Microarray chips were purchased from NimbleGen Systems Inc., Madison, WI. Cells from aerated log-phase cultures (250 ml) (OD600, 0.4 to 0.8) were harvested by centrifugation, RNAs were extracted, and cDNAs were end labeled with biotin. Cells were grown in five different media in triplicate to give a total of 15 RNA samples. The media were LB, M9 plus 15 mM glucose, M9 plus 15 mM succinate, M9 plus 5 mM protocatechuate (PCA), and M9 plus 5 mM PCA plus 15 mM glucose. Hybridization as well as probe intensity analysis was performed by NimbleGen, following company procedures. The custom-made arrays contained 385,298 24-mer oligonucleotide probes targeting sequences within annotated start and end positions of 6,269 annotated S. melilotifeatures (mostly protein coding sequences). The raw data, consisting of probe intensities, were quantile normalized across all experimental replicates. The median intensity of all probes within the annotated region was used as an uncorrected measure of gene expression for each experiment. Background expression was estimated for each experiment by simulating a gene through 10,000 random samples drawn from the normalized intensities of a null probe set of randomly generated sequences. Data for the cbtJKLlocus are presented in Table S1 in the supplemental material and in Fig. 1.
Construction of plasmid integration mutants and reporter gene fusion strains.
To generate promoter fusions to the gfp-lacZreporter genes, a 535-bp DNA fragment upstream of the cbtJopen reading frame (ORF) was PCR amplified using primers P20056F and P20056R and then cloned into the BglII-XhoI sites in pTH1703 to obtain pTH1968 (Fig. 1A). Single-crossover homologous recombination of pTH1968 into the S. melilotigenome resulted in the fusion of the cbtJpromoter region to gfp-lacZand preserved a functional copy of the promoter and all genes at this locus in strain RmP831 (Fig. 1B). To construct gfp-lacZfusions to cbtJand cbtL, internal fragments of cbtJ(543 bp) and cbtL(571 bp) were PCR amplified using the 20056intF and 20056intR primers and the 20058intF and 20058intR primers, respectively, and cloned into the BglII-XhoI sites in pTH1703 to obtain transcriptional fusion plasmids pTH1969 and pTH2030, respectively (Fig. 1A). Cointegration of pTH1969 into the S. melilotigenome generated a fusion of the 5′ end of cbtJto the gfp-lacZgenes and impaired the expression of all downstream genes in this locus in RmP833 (Fig. 1B). However, integration of pTH2030 into S. melilotigenerated a 5′-cbtL::gfp-lacZfusion that maintained functional copies of cbtJand cbtKbut impaired the expression of cbtL, smb20059, and smb20060in strain RmP889 (Fig. 1B). To demonstrate the function of the smb20059and smb20060genes in Co2+uptake, a 486-bp fragment from the 3′ end of cbtLand a 1,201-bp fragment from the 3′ end of smb20059to the 5′ end of smb20060were PCR amplified using the 20058endF and 20058endR primers and the B20059F and ML19261 primers, respectively. These were cloned into the BglII-XhoI sites in pTH1703 to obtain pTH1970 and pTH2270, respectively (Fig. 1A). Integration of plasmid pTH1970 into the S. melilotigenome resulted in strain RmP835, in which the cbtJ, cbtK, and cbtLgenes were functional, whereas the expression of smb20059and smb20060was disrupted (Fig. 1B). Integration of pTH2270 into S. melilotiresulted in a dysfunctional smb20060gene in RmP1485 (smb20060::gfp+-lacZ) (Fig. 1B).
To abolish Cbl biosynthesis in S. meliloti, a cobTmutant was constructed. S. melilotiCobT (SMc00701) shares 91% amino acid sequence identity to the Pseudomonas denitrificansCobT protein, a subunit of the cobalt chelatase complex CobNST, which is required for insertion of cobalt into hydrogenobyrinic acid a,c-diamide in vitamin B12biosynthesis (12). A pTH1522 library fusion plasmid, pFL3283 (8), was restricted with SpeI-XhoI to remove the gfp-lacZreporter genes, filled in by use of Klenow polymerase, and then self-ligated to obtain pTH2293, which carries the 3′ region of cobS(198 bp), the 5′ region of cobT(390 bp), and an intergenic region (50 bp) of the cobSTgenes. Strain RmP1477 (cobT) was generated following integration of pTH2293 into the S. melilotichromosome and was selected on LB medium containing 10 μM AdoCbl. RmP1477 was a Cbl auxotroph and grew only on LB or M9 medium supplemented with AdoCbl.
To construct the replicating reporter plasmid pTH2224, the pTH1703 plasmid (8) was digested with PstI, filled in by use of Klenow polymerase, and then digested with XhoI to obtain a 4,037-bp fragment carrying the gfp-lacZgenes. The BglII site in pTH1919 (Table 1) was deleted by BglII digestion and fill-in by use of Klenow DNA polymerase to obtain pTH2213. A 136-bp Ω terminator from pTH1703 was PCR amplified using primers ML18596 and ML18597 and cloned into the SacI-XbaI sites in pTH2213 to obtain pTH2221. The pTH2221 plasmid was cut with KpnI, filled in by use of Klenow polymerase, cut with XhoI, and then ligated with the gfp-lacZfragment (XhoI-ΔPstI) to yield pTH2224.
To measure transcription from the cbtJpromoter in RmP110 (wild type) and RmP1447 (cobTmutant), a 983-bp fragment upstream from cbtJwas PCR amplified using primers ML18589 and ML18594, cut with SpeI-BamHI, and cloned into the XbaI-BglII sites in pTH2224 to obtain the transcriptional fusion plasmid pTH2237 (see Fig. 6). In order to reveal the role of the B12regulatory element in cbtJexpression, the putative B12riboswitch (see Fig. 4C and 6A) upstream of the cbtJORF was deleted by two-step PCR. A 627-bp fragment upstream of the B12riboswitch and another 221-bp region between the riboswitch and the cbtJORF were PCR amplified using primer pairs ML18589-ML19561 and ML19560-ML18594, respectively. The two products were purified and annealed as the template for a second PCR using primers ML18589 and ML18594. The second PCR product, carrying an 806-bp region lacking the B12riboswitch sequence, was cloned into the SpeI-BamHI sites in pTH2224 to obtain the transcriptional gfp-lacZfusion plasmid pTH2256 (see Fig. 6A). To further analyze the regulatory sequence, a 595-bp region upstream of the transcription start site and a 147-bp region within the 5′-UTR were PCR amplified using primer pairs ML18589-ML18595 and ML18590-ML18594, respectively, and then cloned into the SpeI-BamHI sites in pTH2224 to obtain pTH2238 and pTH2239, respectively (see Fig. 6A). A DNA fragment carrying the 147-bp region upstream of the cbtJORF and the 5′ cbtJcoding region (333 bp) was PCR amplified using primers ML18590 and ML18594 and then inserted into pTH2224 to obtain pTH2240, which was used for DNA sequencing reactions.
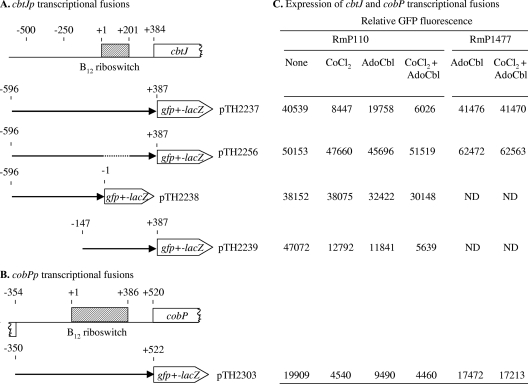
Fig. 6.
Regulation of cbtJand cobPpromoter (cbtJpand cobPp) activities by CoCl2and AdoCbl. (A and B) Transcriptional fusions in S. melilotiRmP110 (wild type) and RmP1477 (cobT) grown in M9-succinate without exogenous CoCl2(none) or with 10 nM CoCl2, 10 μM AdoCbl, or 10 nM CoCl2and 10 μM AdoCbl. (A) The DNA fragments upstream of cbtJwere cloned as transcriptional gfp-lacZfusions in the promoterless, broad-host-range replicating plasmid pTH2224. The location of the transcription start site is marked +1, and the dashed line indicates the deleted B12riboswitch. (B) The region upstream of cobPwas inserted upstream of the gfp-lacZgenes and 5′ of the putative B12riboswitch, designated +1. (C) GFP expression (fluorescence emission [OD600]) given as averages for three independent assays, with SD of <10%. The relative GFP fluorescence was <1,400 units for RmP110 and RmP1477 carrying the empty plasmid pTH2224. ND, not determined.
Fig. 4.
Characterization of the region upstream of cbtJ. The sequences of the cbtJ−35 and −10 promoter recognition sequences and the transcription start site (+1) are indicated. The primers used for mapping the transcription start site are indicated by arrows under the sequences. The sequence of the 201-nt B12riboswitch is shown in bold. The B12box is framed by a dashed-line box. The stem-loop sequences P0/P0′ to P6/P6′ are indicated by arrows above the respective sequences, based on the conserved structure of the B12riboswitch (67). The ribosome binding site (RBS) and start codon of cbtJare shown in bold. The autoradiograph image shows a sequencing gel and the 32P-labeled ML18490 extension products obtained with RNA from RmP110 grown in LB (lane 1). Data obtained with primer ML18489 are not shown but were consistent with those obtained with ML18490. The arrow indicates the primer extension product.
To generate an S. meliloticobPpromoter fusion, an 871-bp intergenic region between cobPand smc04306was PCR amplified using primers ML20074 and ML20075 and inserted upstream of gfp-lacZin pTH2224 to obtain the transcriptional fusion plasmid pTH2303 (see Fig. 6B). β-Galactosidase (LacZ) activity and green fluorescent protein (GFP) were measured as previously described (8).
Co2+uptake assays.
S. melilotiRmP110 (wild type) was grown in LB or LB supplemented with 5 μM CoCl2. Strain RmP833 (cbtJ::gfp+-lacZ) was grown in LB supplemented with 5 μM CoCl2for 16 h, washed three times with 0.85% NaCl, and then subcultured in LB or LB supplemented with 5 μM CoCl2. All cultures for Co2+uptake assays were grown to an OD600of approximately 1. Cells were harvested by centrifugation at 4°C at 4,000 × gfor 20 min, washed three times with transport buffer (50 mM MOPS, pH 7.4, 10 mM MgCl2, and 15 mM succinate), and then resuspended in the buffer to an OD600of about 2 for LB-grown RmP110 and to an OD600of approximately 10 for other strains. Sixty microliters of the cell suspension was added to 510 μl of transport buffer, and following incubation at 30°C for 5 min, assays were initiated through the addition of 30 μl of 10 μM CoCl2labeled with 57Co2+(specific activity, 265 mCi/mmol) to a final concentration of 0.5 μM. Aliquots (0.1 ml) were removed from the assay mixture at different times, immediately passed through nitrocellulose membranes (pore size, 0.45 μm) (HAWP 02500; Millipore, Bedford, MA) that had been presoaked in the same buffer, and immediately washed with the transport buffer (total of 8 ml). The filters were dried and counted using a model 1480 automatic gamma counter (PerkinElmer). All transport assays were performed in triplicate. For chase experiments, a 100-fold excess of unlabeled CoCl2was added to the 57Co2+uptake assay solution at 10 min.
Plant growth and gene expression in nodules.
Alfalfa growth in a nitrogen-deficient growth medium was set up as previously described (1). Plant shoots and nodules were obtained 4 weeks after inoculation. The plant shoots were dried in an oven, and the dry weights were used as an index of symbiotic N2fixation. Preparation of bacteroids from the nodules was carried out as described previously (73), and β-galactosidase activity assays were performed in microtiter plates as described previously (8). β-Galactosidase specific activities were expressed as follows: (A420× 1,000) (time)−1(amount of bacteroid extract)−1(concentration of protein)−1, where time is in minutes, amount of bacteroid extract is in milliliters, and concentration of protein is in mg ml−1.
Biochemicals and radiochemicals.
Cyanocobalamin (CNCbl or vitamin B12), AdoCbl, and CoCl2·6H2O were purchased from Sigma-Aldrich Canada Ltd. 57CoCl2(specific activity, 381 Ci/mmol) was obtained from Amersham Biosciences, GE Healthcare. ICP-MS analysis of the culture media was performed by ActLabs, Ancanster, Ontario, Canada.
Microarray data accession number.
The microarray data are available in the CIBEX database under accession number CBX157.
RESULTS
cbtJKL(smb20056, smb20057, and smb20058) genes are required for growth in LB medium.
The cbtJKLgenes, located on the pSymB megaplasmid of S. meliloti, were originally annotated smb20056, smb20057, and smb20058, respectively (21, 24). These encode a periplasmic substrate-binding protein (CbtJ), a permease (CbtK), and an ATP-binding protein (CbtL) of an ABC-type transport system. As discussed below, these genes were reannotated btuF, btuC, and btuDin the current S. melilotigenome database (4). Based on our data and unpublished work (J. Cheng et al., submitted for publication), we have designated these genes the cobalt transporter genes cbtJ, cbtK, and cbtL, respectively, and we use these names throughout this work. A fourth gene, smb20059, annotated a putative S-adenosylmethionine (SAM)-dependent methyltransferase gene, is located directly downstream of cbtL. The last 2 nucleotides (nt) of smb20059overlap with the insertion sequence element ISRm5(Fig. 1A).
Our interest in the cbtJKLgene cluster arose from the observation that S. melilotiplasmid integration recombinants which are genotypically cbtJ+, cbtK, cbtL, and smb20059negative could not be recovered on LB medium (8) (SmFl3108 in Fig. 1). Microarray experiments using mRNA from wild-type cells showed that the cbtJKLand smb20059genes were highly expressed in cells grown in LB, whereas only background expression was observed in minimal medium (M9) with glucose, succinate, or protocatechuate as a carbon source (see Fig. S1 in the supplemental material). We therefore investigated whether S. meliloticbtJKLmutants could be recovered on M9-succinate medium and found this to be the case. Recombinants RmP831, RmP835 (smb20059negative), and RmP1485, which are all cbtJ+cbtK+cbtL+, were able to grow on both LB and M9-succinate media. However, mutants RmP833, RmFL3108, and RmP889, which are disrupted in one or more genes of the cbtJKLoperon, could not grow on LB, although they could grow on M9-succinate (Fig. 1A). Both the microarray data and the overlapping structure of the cbtJKLgenes strongly suggest that the cbtJKL-smb20059genes are transcribed as an operon. Accordingly, since the integration mutations have polar effects on downstream genes, we have formally established that cbtLis required for growth on LB and that smb20059is not, as strain RmP835 (smb20059mutant) grew on LB.
Co2+is required for growth of cbtJKLmutants under free-living conditions.
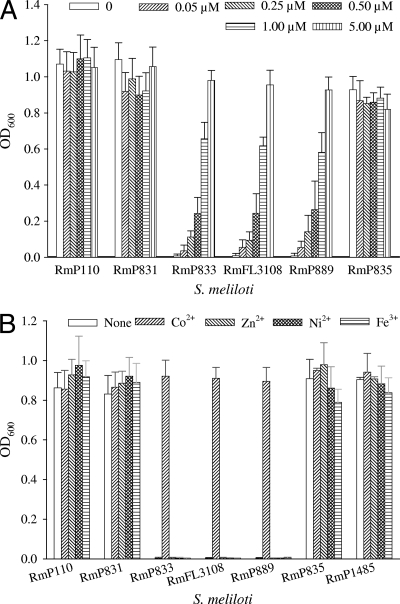
The results suggested that growth of cbtJKLmutant cells in LB required a component present in minimal medium. To identify that component(s), the ingredients of M9 medium, including the trace elements, were added individually to LB, and growth of the cbtJKLmutants was monitored by measuring the OD600. The results from these experiments showed that cbtJKLmutants (RmP833, RmFL3108, and RmP889) grew poorly in LB unless the medium was supplemented with CoCl2(Fig. 2A and data not shown). Growth of the cbtJKLmutants was strongly dependent on the concentration of CoCl2added to the LB medium. At 0.5 μM CoCl2, the mutants grew to an optical density that was approximately 25% that of the wild-type strain RmP110, and growth was essentially rescued with 5 μM CoCl2(Fig. 2A). Strain RmP2361, in which a 50-kb region including the cbtJKLgenes was deleted, grew only in LB medium supplemented with cobalt. Cosmid clones carrying the wild-type cbtJKLgenes allowed this mutant to grow in LB without added cobalt. Because cobalt transport is often linked to the transport of nickel and other metal ions (16), we investigated whether the apparent requirement of the cbtJKLmutants for Co2+could be met by addition of Zn, Ni, or Fe ions. When LB was supplemented with 2 μM ZnSO4, 2 μM NiSO4, or 1 mM FeCl3, none of the metal ions could rescue the growth of S. melilotiRmP833, RmFL3108, or RmP889 (Fig. 2B). These results indicate that the growth defect of the cbtJKLmutants is Co2+specific.
Fig. 2.
Growth of S. melilotiwild-type RmP110 and plasmid integration mutants in LB or LB with Co2+, Zn2+, Ni2+, or Fe3+added. S. melilotistrains were cultured in M9-succinate medium, washed with 0.85% NaCl, and then subcultured into LB. After growing for 16 h, the precultures were inoculated into fresh LB or LB supplemented with CoCl2(0 to 5 μM) (A) or CoCl2(5 μM), ZnSO4(2 μM), NiSO4(2 μM), or FeCl3(1 mM) (B). The OD600was measured after 16 h of incubation.
The M9 and MOPS-buffered minimal media we routinely employ for growth of S. meliloticontain 43 nM CoCl2(70). Both the wild-type RmP110 strain and the cbtJKLmutant strains grew in these minimal media. ICP-MS analysis of the LB medium employed in our experiments detected 100 nM cobalt, a level similar to those previously reported (47). A 100 nM cobalt concentration is >50-fold higher than the 1.7 nM concentration reported to be sufficient to support the growth of S. melilotiin minimal media (31) and over twice the 43 nM CoCl2level we employ in minimal media. It was thus surprising that growth of the cbtJKLmutants in LB required supplementation with cobalt. However, since the yeast extract and tryptone present in LB are strong chelators of metal ions (44), we concluded that there was insufficient bioavailable or free cobalt in LB to support the growth of cbtJKLmutants. Consistent with this suggestion, when a 1 μM concentration of the chelating agent EDTA was added to M9-succinate minimal medium containing 43 nM CoCl2, the cbtJKLdeletion mutant RmP2364 failed to grow, whereas both the wild-type RmP110 strain and the complemented mutant, RmP2364 (ΔcbtJKLstrain plus cbtJKL), grew well. Moreover, in the presence of 1 μM EDTA, increasing the CoCl2concentration to 420 nM allowed the cbtJKLdeletion mutant to grow (see Fig. S2 in the supplemental material).
An smb20056::Tn5mutant of Rm1021 was reported in a collection of mutants of S. melilotiwhose growth in LB was sensitive to a high concentration of salt (350 mM NaCl) (41). Accordingly, we examined the cbtJmutant RmP833 (smb20056::gfp+-lacZ) for growth in LB containing 5 μM CoCl2and found that it grew like the wild-type RmP110 strain, whether or not the medium contained 350 mM NaCl. Thus, under our conditions, the cbtJKLmutants were not sensitive to high salt concentrations. It is possible that the growth differences may reflect differing sources of the LB components.
Growth of a vitamin B12auxotroph and cbtJKLmutants in the presence of AdoCbl.
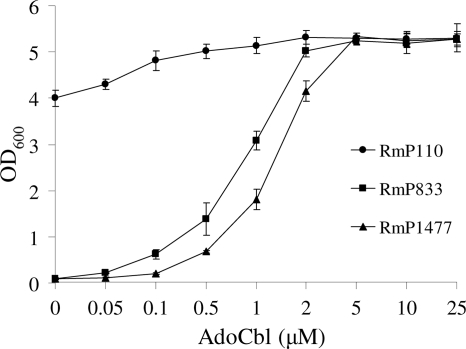
Given the presence of cobalt in vitamin B12and the similarity of CbtJ to the cobalamin-binding protein BtuF, we investigated whether the addition of AdoCbl would allow cbtJKLmutants to grow in LB medium. To interpret these experiments, we determined the concentration of cobalamin that was necessary for growth of an S. melilotiCbl biosynthesis mutant. The data showed that the Cbl biosynthesis mutant RmP1477 (cobT) and the cbtJKLmutant RmP833 had similar growth profiles in LB containing AdoCbl, although the cbtJKLmutant grew more rapidly than the cobTmutant at lower concentrations of AdoCbl (Fig. 3). Thus, while the data suggest that AdoCbl can substitute for cobalt in the cbtJKLmutant, the cbtJKLmutant cells appeared to be unimpaired in the ability to take up AdoCbl. Hence, the CbtJKL system does not appear to transport Cbl.
Fig. 3.
Growth of S. melilotiRmP110 (wild type), RmP833 (cbtJKL), and RmP1477 (cobT) in LB medium supplemented with AdoCbl or left unsupplemented. The strains were initially grown in LB containing 5 μM AdoCbl, washed with 0.85% NaCl, and then inoculated into LB medium supplemented with different concentrations of AdoCbl. The OD600was recorded after incubation for 24 h. Assays were performed in triplicate, and values represent the means ± standard deviations (SD).
The cbtJ5′ region contains a promoter and a vitamin B12riboswitch.
To map the transcriptional start site(s) upstream of cbtJ, we performed primer extension with a primer (ML18489) that overlaps the cbtJATG start codon and a primer (ML18490) 340 nt upstream of the start codon (Fig. 4and data not shown). RNA isolated from LB-grown wild-type cells revealed a transcript that initiated 384 nt upstream of cbtJin both extension reactions (Fig. 4). The deduced −35 and −10 hexanucleotide promoter sequence, 5′-CTTGAC-N17-ATTAAC-3′, showed similarity to the recently derived S. melilotipromoter consensus sequence 5′-CTTGAC-N17-CTATAT-3′, particularly within the more conserved −35 region (38).
A 574-bp DNA sequence upstream of the cbtJORF was analyzed against the Rfam RNA database (http://rfam.sanger.ac.uk/), and a 201-nt region (nt +1 to +201 in Fig. 4) was identified as similar to a conserved RNA structure known as a B12riboswitch (42). A highly conserved sequence called the B12box (70) was located at nucleotides +182 to +192. Characterized B12boxes lie upstream of btuBin E. coli(37) and S. Typhimurium (50) and upstream of the cbiAgene (the first gene in the coboperon) in S. Typhimurium (53). In a survey of conserved RNA structural features associated with genes involved in vitamin B12metabolism and transport in bacteria, Vitreschak et al. (69) identified the same B12riboswitch and B12box upstream of S. meliloticbtJ(smb20056). In that publication and a recent updated annotation of the S. melilotigenome (4), the cbtJ(smb20056), cbtK(smb20057), and cbtL(smb20058) genes were reannotated btuFCD—encoding a vitamin B12transport system. Our data suggest that this system transports cobalt, not vitamin B12, hence the designation cbtJKL.
Repression of cbtJKLexpression by cobalamin and involvement of Co2+in the B12riboswitch.
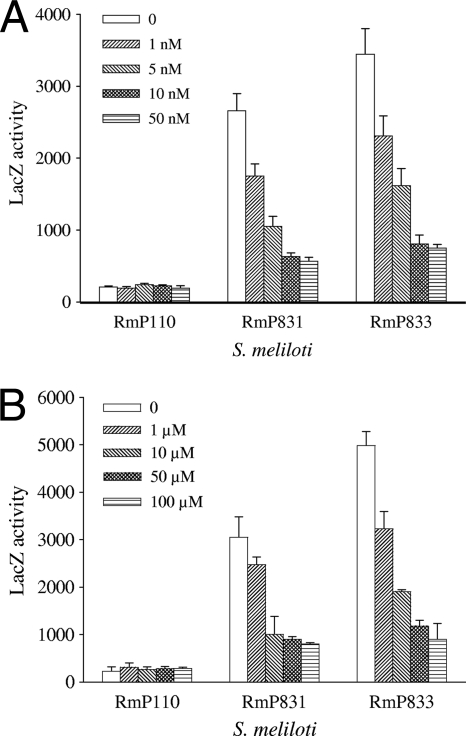
In view of the B12riboswitch-like element identified above and the growth properties of the cbtJKLmutants, the effects of exogenous AdoCbl and CoCl2on cbtJKLexpression were determined. Transcription was examined in the transcriptional gfp-lacZgene fusion strains RmP831 and RmP833, which are phenotypically wild type and mutant, respectively, for the cbtJKLgenes (Fig. 1). In M9-succinate minimal medium, higher levels of cbtJtranscription were found in the cbtJKLmutant than in the wild-type background (Fig. 5). Addition of CoCl2or AdoCbl (or cyanocobalamin [data not shown]) to the medium attenuated cbtJtranscription. We note that cbtJtranscription was much more sensitive to the addition of cobalt than to the addition of AdoCbl. Accordingly, whereas 10 μM AdoCbl reduced cbtJtranscription by approximately 70%, only 10 nM CoCl2was required for a similar reduction in cbtJtranscription (Fig. 5A and B).
Fig. 5.
Nanomolar concentrations of CoCl2and micromolar concentrations of AdoCbl affect cbtJexpression in S. meliloti. Cells were grown in M9-succinate medium, washed with 0.85% NaCl, and then inoculated into M9-succinate with 0 to 50 nM CoCl2(A) or M9-succinate with 0 to 100 μM AdoCbl and no CoCl2(B). After growth for 36 h, LacZ (β-galactosidase) activity was determined. The fusion strains were RmP110 (wild type), RmP831 (cbtJp::gfp-lacZfusion), and RmP833 (cbtJKLmutant with cbtJ::gfp-lacZfusion).
To define cis-acting sequences involved in the modulation of cbtJtranscription by Co2+or Cbl, the full-length promoter region and fragments lacking portions of this region were examined for the ability to drive gfptranscription in a replicating reporter plasmid (pTH2224) (Fig. 6A). In the wild-type strain RmP110, the cbtJpromoter in pTH2237 and pTH2239 initiated transcription to levels that were 40-fold higher than that in cells containing the empty vector pTH2224 (1,400 units) (Fig. 6A). Transcription was repressed upon the addition of 10 nM CoCl2or 10 μM AdoCbl, and this repression was not observed in constructs in which the putative B12riboswitch region (nt +1 to +201 or nt +1 to +387) was deleted in pTH2256 or pTH2238 (Fig. 6A). These data suggested that there are no other regulatory regions (such as binding sites for a transcriptional regulator) upstream of the B12riboswitch.
As a control for the above experiments, we investigated whether Co2+could also modulate the expression of Cbl biosynthesis (cob) genes in S. meliloti. A predicted B12riboswitch and a B12box were previously located at nt −153 to −137 upstream of the cobP(smc04305) start codon (69) (Fig. 4). This region was fused to gfpin plasmid pTH2303, and assays were performed as described for the cbtJpromoter constructs (Fig. 6B). The levels of GFP detected under the various growth conditions demonstrated that the cobPpromoter was negatively regulated by AdoCbl and CoCl2, in a similar fashion to that observed for the cbtJpromoter (Fig. 6A). We infer that this regulation occurs via the B12riboswitch and that Co2+-mediated repression of both the cobPand cbtJpromoters occurs via newly synthesized Co2+-containing Cbl.
To gain further insight into the differential regulatory effects of AdoCbl and CoCl2on the cbtJand cobPpromoters, transcription from these promoters was examined in the cobTmutant RmP1477, in which Cbl biosynthesis was eliminated (Fig. 6A and B). In the cobTmutant, transcription from the cbtJand cobPpromoters (pTH2237 and pTH2303, respectively) was not attenuated upon addition of AdoCbl or CoCl2plus AdoCbl. These results suggest that AdoCbl transport into S. melilotiis inefficient, such that even at an extracellular concentration of 10 μM AdoCbl, the internal concentration of AdoCbl is insufficient to repress expression via the B12riboswitch. In contrast to the case for the wild-type background, the addition of 10 nM cobalt (as in the treatment with CoCl2plus AdoCbl) in the cobTmutant background had no effect on cbtJand cobPtranscription. Thus, both the cbtJand cobPpromoters behaved similarly with respect to cobalt-dependent regulation. These data suggest that cobalt-mediated regulation occurs via newly synthesized Co2+-containing Cbl interacting at the B12riboswitch.
Reduced Co2+accumulation correlates with reduced cbtJKLtranscription.
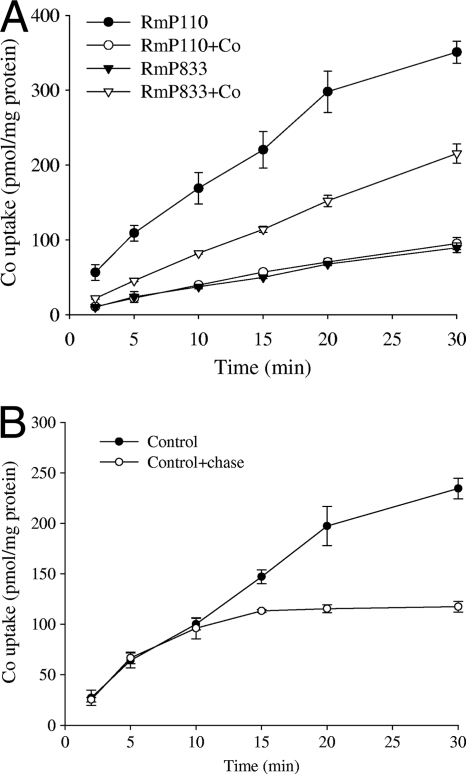
To directly investigate cobalt transport, S. melilotiwild-type (RmP110) and cbtJKLmutant (RmP833) cells were examined for the ability to transport and accumulate 57Co2+. RmP110 cells grown in LB showed the fastest time-dependent Co2+accumulation, and this activity decreased by about 80% in RmP110 cells grown in LB supplemented with 5 μM CoCl2(Fig. 7A). RmP833 cells cultured in LB with 5 μM CoCl2showed 40 to 60% of the Co2+uptake activity of LB-grown RmP110 cells, whereas similar low levels of Co2+transport were observed in RmP833 cultured in LB and RmP110 grown in LB with 5 μM CoCl2. These data suggest that in the cbtJKLmutant (RmP833) cells, an alternate system(s) that can transport Co2+was induced by exogenous Co2+.
Fig. 7.
Cobalt (57Co2+) accumulation by the wild type (RmP110) and a cbtJKLmutant (RmP833) of S. meliloti. Cells were grown in LB or LB supplemented with 5 μM CoCl2, washed, and then resuspended in transport buffer (50 mM MOPS, pH 7.5, 10 mM MgSO4, and 15 mM succinate). Transport assays were initiated by the addition of CoCl2(57Co2+; specific activity, 265 mCi/mmol) to cell suspensions at a final concentration of 0.5 μM. (A) 57Co2+accumulation by RmP110 and RmP833 cells grown in LB or LB with 5 μM CoCl2. (B) 57Co2+accumulation by RmP110 cells grown in LB or with the addition of 50 μM unlabeled CoCl2at 10 min. Each data point represents the average for three independent measurements. The error bars represent SD. Green fluorescence readings (mean emission at 510 nm/OD600± SD) for the cells used in this experiment were 92 ± 11 (RmP110), 80 ± 8 (RmP110 plus CoCl2), 4,927 ± 282 (RmP833), and 1,400 ± 43 (RmP833 plus CoCl2).
To examine the characteristics of 57Co2+uptake, we investigated whether the addition of a 100-fold excess of nonradioactive cobalt would release the 57Co2+accumulated by cells over a 10-min incubation period (Fig. 7B). No release of 57Co2+from the cells was observed, indicating the unidirectional accumulation of 57Co2+by S. meliloti. Moreover, the lack of 57Co2+release from the cells also suggests that nonspecific binding of 57Co2+to the cells did not occur, as we would expect the excess nonradioactive cobalt to displace such nonspecifically bound 57Co2+.
Low cbtJKLexpression in alfalfa nodules.
To investigate cbtJexpression in root nodules and whether the cbtJKLgenes are required for nodule formation and symbiotic N2fixation, various S. melilotistrains were inoculated onto alfalfa seedlings. After growth for 4 weeks under nitrogen-deficient conditions in Leonard jars, plant dry shoot weights were used as an index of N2fixation. The dry shoot weights of plants (averages ± standard errors) inoculated with RmP831, RmP833, and RmP835 (35.1 ± 3.9, 33.1 ± 0.8, and 28.9 ± 4.8 mg/plant, respectively) were similar to that of wild-type RmP110 (32.8 ± 4.6 mg/plant). These results suggest that disruption of the cbtJKLoperon had no effect on symbiotic N2fixation.
To investigate cbtJKLexpression in S. melilotibacteroids, we measured β-galactosidase activity from a cbtJ::gfp-lacZgene fusion and an nifH::gfp-lacZgene fusion in bacteroids from 4-week-old nodules. As expected, the nifHgene, encoding nitrogenase reductase, was highly expressed (mean LacZ specific activity ± standard error, 12,070 ± 1,172), at a level about 110-fold over the RmP110 background level (107 ± 16). The LacZ activities from the cbtJ-cbtL::gfp-lacZfusion genes in RmP831, RmP833, and RmP835 bacteroids were expressed at levels about 2.5-fold above the background level (LacZ specific activity, 280 ± 11, 262 ± 21, and 255 ± 22, respectively). These results suggest that the cbtJKLgenes have little role in symbiotic N2fixation and are expressed only at low levels. It is likely that an alternate transporter(s) is responsible for Co2+uptake into bacteroids, as Co2+plays an essential role in Rhizobium-Medicagosymbiosis (15, 31).
DISCUSSION
Defining pathways for the transport of cobalt into bacteria is of general importance because trace elements, including cobalt, are important modulators of biological processes. Cobalt availability has been shown to limit methane production by methylotrophic methanogens (22) and mercury methylation by the sulfate-reducing bacterium Desulfococcus multivorans(17), and it has been suggested to influence the composition of phytoplankton in the ocean (59). The abundant marine cyanobacterium Prochlorococcusrequires cobalt for growth, and both it and Synechococcusappear to synthesize and excrete cobalt-binding ligands which enhance cobalt assimilation (60).
Here we present evidence that the cbtJKL(smb20056to smb20058) genes encode a cobalt transport system. The CbtJ, CbtK, and CbtL proteins appear to be typical of a solute-binding protein-dependent ABC-type transporter, and this represents one of the first examples of an ABC system for the transport of cobalt. The conclusion that the transported ligand is cobalt is based primarily on the finding that cbtJKLmutants do not grow in LB unless this medium is supplemented with cobalt. This growth requirement appears to be specific for cobalt, as other metals, such as Ni, Zn, and Fe, failed to promote growth (Fig. 2A and B). Moreover, cbtJKLexpression was downregulated in response to the addition of exogenous cobalt (Fig. 5A), and 57Co2+accumulation was correlated with cbtJKLexpression (Fig. 7). In another report, we also demonstrate that the solute binding protein CbtJ can bind to cobalt (Cheng et al., submitted).
Other cobalt transport systems have been described (16, 33, 56, 57, 74), and frequently these systems contain B12riboswitches, as is the case for the cbtJKLgenes described here. Recently, an ABC transporter (FecDE and CeuE) in Helicobacter mustelaewas reported to contribute to nickel and cobalt acquisition. The authors concluded that this transporter was not specific for nickel, as a fecDmutant showed reduced cellular cobalt levels and increased cobalt resistance (65).
Another uptake system(s) present in S. melilotican also transport cobalt, as the growth of cbtJKLmutants in LB supplemented with cobalt requires its uptake via another system(s). A growth phenotype similar to that of the cbtJKLmutants was reported for a Ralstonia eutropha hoxNmutant defective in a high-affinity, nickel-specific permease. The ability of this hoxNmutant to grow with H2as an energy source was restored by increasing the concentration of nickel in the medium (13). While the identity of the alternate S. melilotitransporter(s) remains to be determined, one candidate is the Mg2+transporter CorA (28, 64). It is unclear whether its affinity for Co would be physiologically sufficient, although the cellular cobalt requirements are presumably very low. Interestingly, in the cbtJKLbackground, the addition of cobalt to the medium appeared to induce the alternate cobalt uptake system(s) (Fig. 7). S. melilotidoes not have homologs of the CbiMNQO or NiCoT cobalt transporters (56, 74). However, a gene (smb20556) designated cbtC, which has an upstream B12riboswitch region, has been suggested to encode a putative cobalt transport protein (55). While we have not directly examined smb20556for a role in cobalt uptake, in other work we observed that deletion of the smb20556gene region had no effect on growth in LB or minimal medium (data not shown).
The obvious inability of the cbtJKLmutants to grow in LB medium contrasts with the growth phenotype observed in minimal medium, where a clear growth phenotype was observed only upon addition of a chelating agent. Strikingly, nanomolar quantities of cobalt restored growth in minimal medium, whereas micromolar quantities were required for growth of cbtJKLmutants in LB. This clear phenotypic difference appears to result from the chelation/binding of cobalt by the yeast extract and tryptone components present in LB. These bind tightly to metal ions, and this dramatically affects their availability to cells (29). For example, Ramamoorthy and Kushner (49) detected no free copper in a nutrient broth solution (0.3% beef extract and 0.5% peptone) containing 3 mM copper. While such bioavailability effects are generally considered with respect to the toxicity of metal ions, here the metal binding influenced the availability of cobalt as a nutrient. Thus, while ICP-MS analysis detected 100 nM Co2+in LB, the actual concentration of free cobalt available to the cells must be much less than the 2 nM concentration required for growth of cbtJKLmutants in minimal media (36, 70).
Growth experiments suggest that the CbtJKL system does not transport cobalamins. Thus, while either cobalt or AdoCbl restored growth to cbtJKLmutants on LB, the concentrations of AdoCbl required for growth of the cbtJKLmutants were similar to the AdoCbl concentrations required to grow a cobalamin biosynthesis mutant (Fig. 3). We note that the cbtJKLgenes were highly expressed in the cobTmutant (Fig. 6), but in both the cobTand cbtJKLmutants, it appears that cobalamin was transported by a low-affinity system. In contrast, E. coli, S.Typhimurium, and Halobacteriumpossess high-affinity cobalamin transport systems, and the addition of 1 nM (or less) cobalamin is sufficient to allow cobalamin auxotrophs to grow like the wild-type strains (6, 68, 72). The absence of a high-affinity cobalamin transport system in S. melilotisuggests that the soil environment in which S. melilotilives lacks sufficient cobalamin or incomplete corrinoids to support the cost of carrying these accessory uptake genes. However, the related alphaproteobacterium Rhodobacter sphaeroidesstrain 2.4.1 possesses a BtuBFCD-like high-affinity cobalamin transporter (RSP_2402 to RSP_2405) (26). Interestingly, this strain also appears to possess cbtJKLhomologues (RSP_3392 to RSP_3390) with a B12riboswitch in the predicted 5′ region (Cheng et al., submitted).
Transcription of the cbtJKLgenes increased upon cobalt depletion, and our data suggest that this regulation occurred via a B12riboswitch located in the mRNA 5′ of cbtJ. The data are consistent with a model whereby cobalt-loaded vitamin B12interacts at the B12riboswitch and terminates cbtJKLtranscription. Gallo et al. (24) have shown that the corrin ring plays a crucial role in the switching structure of the btuBriboswitch. Data obtained with transcriptional and translational reporter fusions (Fig. 6and data not shown) suggest that the cbtJriboswitch functions via termination of transcription. However, more detailed in vitroand in vivoanalyses of this region are required to demonstrate a precise mechanism. The presence of free metal ions in the cytoplasm is toxic to the cell (47). In the case of noncorrin enzymes such as nitrile hydratase, the transported cobalt is bound to a chaperon prior to its insertion into the protein (75). Upon entry to the cytoplasm via the CbtJKL transporter, we assume that cobalt is rapidly incorporated into B12.
In previous reports based on informatics, the smb20056, smb20057, and smb20058genes were designated btuFCDgenes encoding a vitamin B12transport system (4, 69). In another report, on the basis of mRNA transcript nucleotide sequencing, Mao et al. (39) suggested that a small, 180-nucleotide open reading frame (vbismb0078) is located in the 5′-UTR of cbtJ. As discussed above, our experimental data suggest that this region contains a B12riboswitch. We note that the transcribed region upstream of cbtJis 384 nucleotides in length and that the B12riboswitch spans a 201-nt region (nt +1 to +201) (Fig. 4). The 384 nucleotides are considerably longer than the equivalent B12riboswitch-containing regions upstream of the btuBgenes in E. coliand S.Typhimurium (37, 50). It is therefore possible that additional regulatory elements may be present. In this respect, it is interesting that an AraC-type transcription regulator gene, smb20055, lies upstream of cbtJ; however, disruption of this gene had no effect on cbtJexpression (data not shown).
S. melilotiis known to require micronutrient concentrations of cobalt for growth (31, 36), and the ribonucleotide reductase and methionine synthase enzymes from S. melilotiuse vitamin B12as a coenzyme (9, 30). In the early 1960s, trace element concentrations of cobalt were shown to be required for symbiotic N2fixation (15). Cobalamin biosynthesis is required for symbiotic N2fixation (66), and as expected, the cobTmutant (RmP1477) formed Fix−nodules on alfalfa (data not shown). Moreover, Taga and Walker (66) recently showed that the cobalamin-dependent ribonucleotide reductase encoded by nrdJis required for symbiotic N2fixation, as a cobalamin-independent ribonucleotide reductase failed to restore symbiotic N2fixation to an nrdJmutant of S. meliloti. Under the plant growth conditions employed in this study, the cbtJKLgenes were expressed at low levels in N2-fixing bacteroids, and thus the intracellular cobalt levels in bacteroids appeared to be sufficient to repress cbtJKLexpression. Indeed, the Fix+phenotype of cbtJKLmutants demonstrates that sufficient cobalt for symbiotic N2fixation is taken up by an alternate transporter(s) in bacteroids. To demonstrate a cobalt requirement for symbiotic N2fixation, it was necessary for Delwiche et al. to explicitly remove cobalt from all components of the plant nutrient solution (15). It remains to be determined whether such plant growth conditions will reveal a symbiotic role for the cbtJKLsystem.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rahat Zaheer for primer extension analyses and Catharine White and Allyson MacLean for comments and suggestions on this work. We thank Christopher Wood for use of a model 1480 automatic gamma counter and anonymous reviewers for helpful comments.
This work was supported with funding from NSERC (Canada), from Genome Canada through the Ontario Genomics Institute, and from the Ontario Research and Development Challenge Fundto T.M.F.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Bardin S., Dan S., Osteras M., Finan T. M. 1996. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 178:4540–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bassford P. J., Jr., Bradbeer C., Kadner R. J., Schnaitman C. A. 1976. Transport of vitamin B12in tonBmutants of Escherichia coli. J. Bacteriol. 128:242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reference deleted.
- 4. Becker A., et al. 2009. A portal for rhizobial genomes: RhizoGATE integrates a Sinorhizobium melilotigenome annotation update with postgenome data. J. Biotechnol. 140:45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borths E. L., Locher K. P., Lee A. T., Rees D. C. 2002. The structure of Escherichia coliBtuF and binding to its cognate ATP binding cassette transporter. Proc. Natl. Acad. Sci. U. S. A. 99:16642–16647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cadieux N., et al. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184:706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campbell G. R. O., et al. 2006. Sinorhizobium melilotibluBis necessary for production of 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc. Natl. Acad. Sci. U. S. A. 103:4634–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cowie A., et al. 2006. An integrated approach to functional genomics: construction of a novel reporter gene fusion library for Sinorhizobium meliloti. Appl. Environ. Microbiol. 72:7156–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cowles J. R., Evans H. J., Russell S. A. 1969. B12coenzyme-dependent ribonucleotide reductase in Rhizobiumspecies and the effects of cobalt deficiency on the activity of the enzyme. J. Bacteriol. 97:1460–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cowles J. R., Evans H. J. 1968. Some properties of the ribonucleotide reductase from Rhizobium meliloti. Arch. Biochem. Biophys. 127:770–778 [DOI] [PubMed] [Google Scholar]
- 11. Davidson A. L., Chen J. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241–268 [DOI] [PubMed] [Google Scholar]
- 12. Debussche L., et al. 1992. Assay, purification, and characterization of cobaltochelatase, a unique complex enzyme catalyzing cobalt insertion in hydrogenobyrinic acid a,c-diamide during coenzyme B12biosynthesis in Pseudomonas denitrificans. J. Bacteriol. 174:7445–7451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Degen O., Kobayashi M., Shimizu S., Eitinger T. 1999. Selective transport of divalent cations by transition metal permeases: the Alcaligenes eutrophusHoxN and the Rhodococcus rhodochrousNhlF. Arch. Microbiol. 171:139–145 [DOI] [PubMed] [Google Scholar]
- 14. De Hertogh A. A., Patrica A. M., Evans H. J. 1964. Effect of cobalt on the oxidation of propionate by Rhizobium meliloti. J. Bacteriol. 87:746–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delwiche C. C., Johnson C. M., Reisenauer H. M. 1961. Influence of cobalt on nitrogen fixation by Medicago. Plant Physiol. 36:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eitinger T., Suhr J., Moore L., Smith J. A. 2005. Secondary transporters for nickel and cobalt ions: theme and variations. Biometals 18:399–405 [DOI] [PubMed] [Google Scholar]
- 17. Ekstrom E. B., Morel F. M. 2008. Cobalt limitation of growth and mercury methylation in sulfate-reducing bacteria. Environ. Sci. Technol. 42:93–99 [DOI] [PubMed] [Google Scholar]
- 18. Entcheva P., Phillips D. A., Streit W. R. 2002. Functional analysis of Sinorhizobium melilotigenes involved in biotin synthesis and transport. Appl. Environ. Microbiol. 68:2843–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Escalante-Semerena J. C. 2007. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J. Bacteriol. 189:4555–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Finan T. M., Kunkel B., Devos G. F., Signer E. R. 1986. Second symbiotic megaplasmid in Rhizobium meliloticarrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finan T. M., et al. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. U. S. A. 98:9889–9894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Florencio L., Field A. J., Lettinga G. 1994. The importance of cobalt for individual trophic groups in an anaerobic methanol-degrading consortium. Appl. Environ. Microbiol. 60:227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galibert F., et al. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672 [DOI] [PubMed] [Google Scholar]
- 24. Gallo S., Mundwiler S., Alberto R., Sigel R. K. 2011. The change of corrin-amides to carboxylates leads to altered structures of the B12-responding btuB riboswitch. Chem. Commun. (Cambridge) 47:403–405 [DOI] [PubMed] [Google Scholar]
- 25. Grass G., et al. 2005. The metal permease ZupT from Escherichia coliis a transporter with a broad substrate spectrum. J. Bacteriol. 187:1604–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gray M. J., Tavares N. K., Escalante-Semerena J. C. 2008. The genome of Rhodobacter sphaeroidesstrain 2.4.1 encodes functional cobinamide salvaging systems of archaeal and bacterial origins. Mol. Microbiol. 70:824–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hebbeln P., Eitinger T. 2004. Heterologous production and characterization of bacterial nickel/cobalt permeases. FEMS Microbiol. Lett. 230:129–135 [DOI] [PubMed] [Google Scholar]
- 28. Hmiel S. P., Snavely M. D., Miller C. G., Maguire M. E. 1986. Magnesium transport in Salmonella typhimurium: characterization of magnesium influx and cloning of a transport gene. J. Bacteriol. 168:1444–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hughes M., Poole R. K. 1991. Metal speciation and microbial growth—the hard (and soft) facts. J. Gen. Microbiol. 137:725–734 [Google Scholar]
- 30. Inukai S., Sato K., Shimizu S. 1977. Relationship of cobalt requirement to vitamin B12-dependent methionine synthesis in Rhizobium meliloti. Agric. Biol. Chem. 41:2229–2234 [Google Scholar]
- 31. Kliewer M., Evans H. J. 1962. Effect of cobalt deficiency on the B12coenzyme content of Rhizobium meliloti. Arch. Biochem. Biophys. 97:427–429 [DOI] [PubMed] [Google Scholar]
- 32. Kobayashi M., Shimizu S. 1999. Cobalt proteins. Eur. J. Biochem. 261:1–9 [DOI] [PubMed] [Google Scholar]
- 33. Komeda H., Kobayashi M., Shimizu S. 1997. A novel transporter involved in cobalt uptake. Proc. Natl. Acad. Sci. U. S. A. 94:36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Köster W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152:291–301 [DOI] [PubMed] [Google Scholar]
- 35. Reference deleted.
- 36. Lowe R. H., Evans H. J. 1962. Cobalt requirement for the growth of rhizobia. J. Bacteriol. 83:210–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lundrigan M. D., Koster W., Kadner R. J. 1991. Transcribed sequences of the Escherichia colibtuBgene control its expression and regulation by vitamin B12. Proc. Natl. Acad. Sci. U. S. A. 88:1479–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. MacLellan S. R., MacLean A. M., Finan T. M. 2006. Promoter prediction in the rhizobia. Microbiology 152:1751–1763 [DOI] [PubMed] [Google Scholar]
- 39. Mao C., Evans C., Jensen R., Sobral B. 2008. Identification of new genes in Sinorhizobium melilotiusing the Genome Sequencer FLX system. BMC Microbiol. 8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martens H., Barg H., Warren M., Jahn D. 2002. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58:275–285 [DOI] [PubMed] [Google Scholar]
- 41. Miller-Williams M., Loewen P. C., Oresnik I. J. 2006. Isolation of salt-sensitive mutants of Sinorhizobium melilotistrain Rm1021. Microbiology 152:2049–2059 [DOI] [PubMed] [Google Scholar]
- 42. Nahvi A., Barrick J. E., Breaker R. R. 2004. Coenzyme B12riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 32:143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nies D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313–339 [DOI] [PubMed] [Google Scholar]
- 44. Nou X., Kadner R. J. 1998. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuBexpression in Escherichia coli. J. Bacteriol. 180:6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nou X., Kadner R. J. 2000. Adenosylcobalamin inhibits ribosome binding to btuBRNA. Proc. Natl. Acad. Sci. U. S. A. 97:7190–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reference deleted.
- 47. Outten C. E., O'Halloran T. V. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492 [DOI] [PubMed] [Google Scholar]
- 48. Pinedo C. A., Bringhurst R. M., Gage D. J. 2008. Sinorhizobium melilotimutants lacking PTS enzymes HPr or EIIA are altered in diverse processes including carbon metabolism, cobalt requirements and succinoglycan production. J. Bacteriol. 190:2947–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramamoorthy S., Kushner D. J. 1975. Binding of mercuric and other heavy metal ions by microbial growth media. Microb. Ecol. 2:162–176 [DOI] [PubMed] [Google Scholar]
- 50. Ravnum S., Andersson D. I. 1997. Vitamin B12repression of the btuBgene in Salmonella typhimuriumis mediated via a translational control which requires leader and coding sequences. Mol. Microbiol. 23:35–42 [DOI] [PubMed] [Google Scholar]
- 51. Ravnum S., Andersson D. I. 2001. An adenosyl-cobalamin (coenzyme-B12)-repressed translational enhancer in the cobmRNA of Salmonella typhimurium. Mol. Microbiol. 39:1585–1594 [DOI] [PubMed] [Google Scholar]
- 52. Richter-Dahlfors A. A., Ravnum S., Andersson D. I. 1994. Vitamin B12repression of the coboperon in Salmonella typhimurium: translational control of the cbiAgene. Mol. Microbiol. 13:541–553 [DOI] [PubMed] [Google Scholar]
- 53. Richter-Dahlfors A. A., Andersson D. I. 1992. Cobalamin (vitamin B12) repression of the coboperon in Salmonella typhimuriumrequires sequences within the leader and the 1st translated open reading frame. Mol. Microbiol. 6:743–749 [DOI] [PubMed] [Google Scholar]
- 54. Rioux C., Kadner R. 1989. Vitamin B12transport in Escherichia coliK12 does not require the btuEgene of the btuCEDoperon. Mol. Gen. Genet. 217:301–308 [DOI] [PubMed] [Google Scholar]
- 55. Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. 2003. Comparative genomics of the vitamin B12metabolism and regulation in prokaryotes. J. Biol. Chem. 278:41148–41159 [DOI] [PubMed] [Google Scholar]
- 56. Rodionov D. A., Hebbeln P., Gelfand M. S., Eitinger T. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188:317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rodionov D. A., et al. 2009. A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 191:42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Roth J. R., Lawrence J. G., Rubenfield M., Kiefferhiggins S., Church G. M. 1993. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 175:3303–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saito M. A., Moffett J. W., Chisholm S. W., Waterbury J. 2002. Cobalt limitation and uptake in Prochlorococcus. Limnol. Oceanogr. 47:1629–1636 [Google Scholar]
- 60. Saito M. A., Rocap G., Moffett J. W. 2005. Production of cobalt binding ligands in a Synechococcusfeature at the Costa Rica upwelling dome. Limnol. Oceanogr. 50:279–290 [Google Scholar]
- 61. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 62. Sato K., Inukai S., Shimizu S. 1974. Vitamin B12-dependent methionine synthesis in Rhizobium meliloti. Biochem. Biophys. Res. Commun. 60:723–728 [DOI] [PubMed] [Google Scholar]
- 63. Schmidt T., Schlegel H. G. 1994. Combined nickel-cobalt-cadmium resistance encoded by the ncclocus of Alcaligenes xylosoxidans31A. J. Bacteriol. 176:7045–7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith R. L., Banks J. L., Snavely M. D., Maguire M. E. 1993. Sequence and topology of the CorA magnesium transport systems of Salmonella typhimuriumand Escherichia coli. Identification of a new class of transport protein. J. Biol. Chem. 268:14071–14080 [PubMed] [Google Scholar]
- 65. Stoof J., Kuipers E. J., Klaver G., van Vliet A. H. 2010. An ABC transporter and a TonB ortholog contribute to Helicobacter mustelaenickel and cobalt acquisition. Infect. Immun. 78:4261–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Taga M. E., Walker G. C. 2010. Sinorhizobium melilotirequires a cobalamin-dependent ribonucleotide reductase for symbiosis with its plant host. Mol. Plant Microbe Interact. 23:1643–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Taga M. E., Larsen N. A., Howard-Jones A. R., Walsh C. T., Walker G. C. 2007. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446:449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Van Bibber M., Bradbeer C., Clark N., Roth J. R. 1999. A new class of cobalamin transport mutants (btuF) provides genetic evidence for a periplasmic binding protein in Salmonella typhimurium. J. Bacteriol. 181:5539–5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vitreschak A. G., Rodionov D. A., Mironov A. A., Gelfand M. S. 2003. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9:1084–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Watson R. J., Heys R., Martin T., Savard M. 2001. Sinorhizobium meliloticells require biotin and either cobalt or methionine for growth. Appl. Environ. Microbiol. 67:3767–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Winkler W. C., Breaker R. R. 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59:487–517 [DOI] [PubMed] [Google Scholar]
- 72. Woodson J. D., Reynolds A. A., Escalante-Semerena J. C. 2005. ABC transporter for corrinoids in Halobacteriumsp. strain NRC-1. J. Bacteriol. 187:5901–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yuan Z. C., Zaheer R., Finan T. M. 2006. Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 188:1089–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang Y., Rodionov D. A., Gelfand M. S., Gladyshev V. N. 2009. Comparative genomic analyses of nickel, cobalt and vitamin B12utilization. BMC Genomics 10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou Z., Hashimoto Y., Kobayashi M. 2009. Self-subunit swapping chaperone needed for the maturation of multimeric metalloenzyme nitrile hydratase by a subunit exchange mechanism also carries out the oxidation of the metal ligand cysteine residues and insertion of cobalt. J. Biol. Chem. 284:14930–14938 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.