Fig. 12.
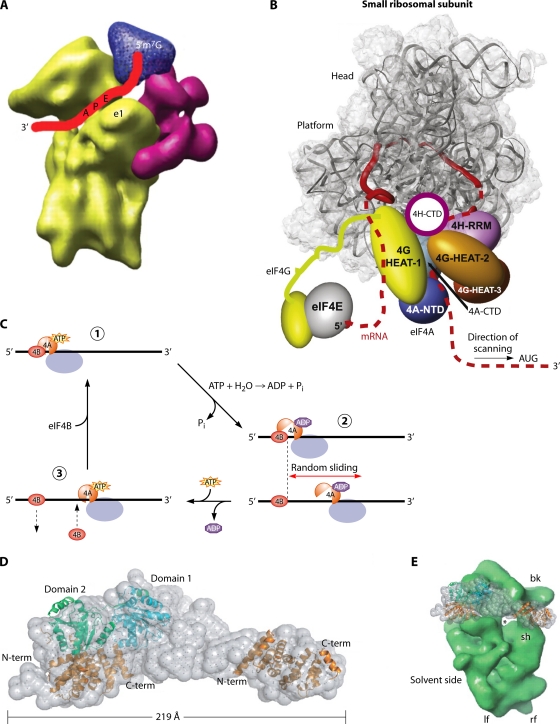
Models of the scanning PIC. (A) Cryo-EM model of mammalian eIF3 (magenta) and eIF4G (blue) binding to the 40S subunit (yellow), annotated to depict the path of the mRNA (red ribbon) in the decoding center and exit channel (Reproduced from reference 194 with permission from AAAS.) (B) Model for the scanning PIC in which the eIF4G N-terminal region remains bound to the cap-eIF4E assembly and the HEAT-1 domain of eIF4G interacts with mRNA sequences immediately 5′ of the 40S exit channel, causing the mRNA to form a loop between the cap and HEAT-1 that grows in size as scanning proceeds. eIF4A, bound to HEAT-1, interacts with mRNA sequences located 3′ of the PIC to destabilize the secondary structure before it reaches the 40S subunit, and eIF4H prevents the reannealing of the unwound, single-stranded nucleotides before they enter the entry channel pore. (Reproduced from reference 134 with permission from Elsevier.) (C) A Brownian ratchet mechanism of scanning in which eIF4A undergoes cycles of binding and dissociation from the mRNA, driven by ATP binding and hydrolysis, near the mRNA exit channel (where it is positioned by eIF4G). eIF4B interacts with the eIF4A·ATP·mRNA complex but dissociates from the mRNA more slowly than does eIF4A·ADP, allowing it to act as a pawl to prevent 3′-to-5′ backsliding. Thus, the random sliding of the ribosome can occur only in the 3′ direction and is fixed at the new location by the next cycle of eIF4A·ATP·mRNA·eIF4B complex assembly. (Reproduced from reference 199 with permission of the publisher [copyright 2009 American Chemical Society].) (D, left) Docking of the crystal structure of yeast eIF4A to eIF4G and the HEAT-1 domain of human eIF4G1 into the SAXS envelope of the complex between eIF4A and the eIF4G fragment containing HEAT domains 1 to 3. (Right) Comparison of the sizes of the 40S subunit and the eIF4A-eIF4G complex. (Reproduced from reference 149 by permission of Oxford University Press.)