Abstract
Summary: Bacteriophages belonging to the order Caudovirales possess a tail acting as a molecular nanomachine used during infection to recognize the host cell wall, attach to it, pierce it, and ensure the high-efficiency delivery of the genomic DNA to the host cytoplasm. In this review, we provide a comprehensive analysis of the various proteins constituting tailed bacteriophages from a structural viewpoint. To this end, we had in mind to pinpoint the resemblances within and between functional modules such as capsid/tail connectors, the tails themselves, or the tail distal host recognition devices, termed baseplates. This comparison has been extended to bacterial machineries embedded in the cell wall, for which shared molecular homology with phages has been recently revealed. This is the case for the type VI secretion system (T6SS), an inverted phage tail at the bacterial surface, or bacteriocins. Gathering all these data, we propose that a unique ancestral protein fold may have given rise to a large number of bacteriophage modules as well as to some related bacterial machinery components.
INTRODUCTION
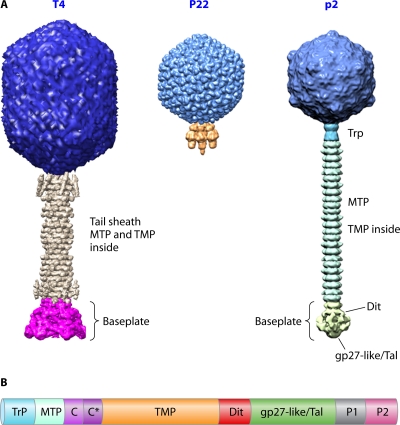
Bacterial viruses (phages or bacteriophages) are very efficient nanomachines designed to infect their hosts with exquisite specificity and efficacy. The vast majority of them belong to the order Caudovirales and possess a double-stranded DNA (dsDNA) genome enclosed in a polyhedral head, being most frequently icosahedral, to which a tail is attached. They are the most numerous biological entity on earth, with an estimated number of 1031 tailed phages in the biosphere (11, 31). They are arguably very ancient as a group, with some estimates placing their ancestors before the divergence of the Bacteria from the Archaea and Eukarya (11). The bacteriophage tail is a molecular machine used during infection to recognize the host and ensure efficient genome delivery to the cell cytoplasm. Its morphology serves as a basis for the classification of Caudovirales phages into three distinct families (Fig. 1A): the Myoviridae, possessing a complex contractile tail (e.g., T4, Mu, or ϕKZ); the Podoviridae, bearing a short noncontractile tail (e.g., P22, ϕ29, or T7); and the Siphoviridae, characterized by their long noncontractile tail (e.g., λ, SPP1, HK97, or lactococcal phages).
Fig. 1.
(A) The three Caudovirales families. From left to right are the Myoviridae (T4), the Podoviridae (P22), and the Siphoviridae (p2). (B) Schematic representation of the typical genome organization within the Siphoviridae tail morphogenesis module (this organization is also observed for several myophages with some adaptations). Trp, tail terminator; MTP, major tail protein; C and C*, tail chaperones; TMP, tape measure protein; Dit, distal tail protein; gp27-like/Tal (tail-associated lysozyme or tail fiber), the presence of a C-terminal domain depends on the phage considered; P1 and P2, baseplate/tip peripheral proteins (their number varies among phages).
The dramatic divergence of bacteriophage genomes is an obstacle that frequently prevents the detection of homology between proteins and, thus, the determination of phylogenetic links between phages. For instance, sequence similarity between Siphoviridae major tail proteins (MTPs), which have been experimentally demonstrated to form the phage tail tube, is often not detectable (74). However, the high degree of conservation of function-associated gene orders in regions encoding morphogenesis modules is striking when numerous phage genomes belonging to the Siphoviridae, Podoviridae, and Myoviridae families are compared (15, 20, 72, 86). This feature is especially interesting because it allows the accurate identification of protein functions in totally unknown phages, provided that genomes are sequenced, even in the absence of detectable sequence similarity with characterized genes. A comparison of the P22 (Podoviridae) and λ (Siphoviridae) genetic maps achieved 3 decades ago demonstrated a similar organization, in which regions without sequence similarity are interspersed with regions exhibiting similarity (9). It is noteworthy that the conserved segments are often found in regions without a known function, as if regions being relatively free to diverge were constrained to be flanked by conserved ones.
Another strategy to infer evolutionary connections between different proteins with similar functions relies on the comparison of their structures (10). Indeed, it was shown at an early stage that protein sequences diverge faster than their structures. Therefore, in the case of a shared architecture between two or more proteins, one can assume that a common ancestor might be at the origin of such proteins. With the advent of structural genomics and the progresses in protein structure determinations, the structural comparison approach becomes a valuable and efficient tool to detect evolutionary links.
In this review, we provide a comprehensive analysis of the various structural proteins constituting tailed bacteriophages from a structural viewpoint, with the goal of pinpointing the resemblances within and between functional modules such as connectors, tails, or baseplates (the packaging machinery components will not be covered here, as they do not belong to phage structural proteins). We also extend this comparison to bacterial machineries for which shared molecular homology with phages has been recently revealed, as in the case of the type VI secretion system (T6SS) or bacteriocins (45, 63, 91). Interestingly, the conservation appears to extend beyond the bacterial kingdom, as some observed folds are shared with eukaryotic viruses, which place them at the origin of life (43, 44, 63, 78, 91). Gathering all these data, we propose that a unique ancestral protein fold has given rise to a large number of bacteriophage modules as well as to some related components found in cell wall-embedded bacterial nanomachines.
TAILED BACTERIOPHAGES
The Modular Nature of Phage Genomes
As stated above, phage genetic maps are characterized by similar organizations based on the alternation of regions with and those without sequence similarity (9). A typical example is the receptor-binding/receptor-blocking module observed for several members of the Siphoviridae infecting Gram-negative bacteria, such as those of phages T5 and BF23 (59). Indeed, no sequence similarity is observed, at the nucleotidic level, between the two receptor-binding protein (RBP)/lipoprotein (Llp) couples formed by oad-llpT5 and hrs-llpBF23, with each one being arranged as an inverted tandem (59). This organization prevents the separation of the two components present within a given module, as they must bind to the same outer membrane receptor to be efficient. Hence, this module has to be exchanged as a whole to confer a selective advantage to the phage into which it will be introduced.
A more detailed illustration is provided by the structural gene module encoding tail components in most members of the Siphoviridae, including phage λ (infecting a Gram-negative host) and phage TP901-1/SPP1 (infecting Gram-positive bacteria), which is found at an equivalent location in the genomes of all characterized members of this phage family. Moreover, a canonical scheme of gene organization within this module has been observed and can be described by the following consecutive open reading frame (ORF) order: the tail terminator, the MTP, the two chaperones (with a conserved programmed translational frameshift [90]), the tape measure protein (TMP), the baseplate hub (Dit), the tail fiber, and baseplate/tip peripheral proteins (Fig. 1B). This organization has also been observed for several myophages, with some adaptations due (at least in part) to the distinct morphological tail aspect of this phage family (69). According to the principle of parsimony, the most probable explanation for this observation is an evolutionary connection between bacteriophages (or at least between all long-tailed structures) and can be interpreted as an argument in favor of a common origin for all long-tailed phages. As the evolution of phages was proposed to involve the exchange of functional modules via a loss or acquisition of genetic material by recombination between phages and also between phages and their hosts (as well as with prophages) (9, 31, 58), this generic gene arrangement might be a facilitator of genetic brewing to assist bacteriophages in their permanent adaptation to changing environmental conditions or in the quest of infecting new hosts. Therefore, a major advantage of modular evolution is probably to provide virions with easy access to a large array of functional specificities by means of homologous recombination. Evolution should thus be considered to be acting on functional modules rather than on viruses themselves, and those modules can be of variable sizes, such as a morphogenesis gene block, a single gene, or a protein domain-encoding sequence.
Limited Number of Folds for Phage Structural Proteins
Extensive structural studies have been carried out on bacteriophages during the last decade, yielding a myriad of structures for several virion components from the Siphoviridae, Podoviridae, and Myoviridae families. This therefore provides a strong basis to perform thorough comparisons among different phage building blocks and study the evolutionary trajectories of these proteins. These data allowed us to observe that virtually all proteins constituting the tail organelle and host adsorption apparatus core of long-tailed phages share striking structural similarities.
The capsid subunit.
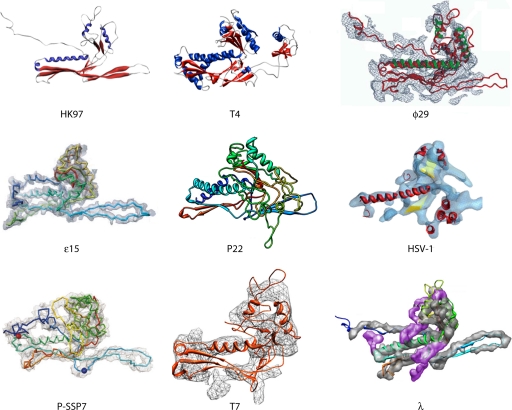
The HK97 major capsid protein (gp5*) fold was demonstrated to be the archetype capsid protein fold in the Caudovirales, including members of the Siphoviridae (24, 25, 46), the Myoviridae (26), and the Podoviridae (1, 16, 33, 34, 53, 60), but also in distantly related virions such as herpes simplex viruses (4) as well as in encapsulins that form a sequestrated icosahedral environment in bacterial physiology (82). Even in a complex capsid such as that of myophage T4, the two different proteins forming the icosahedral shell (gp23* and gp24*) share a similar folding pattern with each other (and are thus most probably paralogous) as well as with all other tailed phages (hence involving an orthologous relationship) (26). Clearly, an ancestral protein module was at the origin of all these icosahedral structures, therefore establishing a lineage among extremely diverse biological systems (Fig. 2). Considering the remarkable abundance of tailed phages, it is likely that the HK97-like gp5* represents the absolute most abundant fold in the biosphere. It is worth mentioning that in addition to the conserved architecture of the capsid building blocks found in the above-mentioned virions, a comparable mechanistic strategy appears to govern the maturation pathway leading from initially assembled fragile proheads to final mature and robust heads, which can sustain significant internal pressure due to the presence of the densely packaged genome.
Fig. 2.
The HK97 lineage. Shown is a gallery of HK97-like capsid protein structures determined by using crystallography for HK97 gp5 and T4 gp24 or cryo-EM for ϕ29 gp5, ε15 gp7, P22 gp5, herpes simplex virus type 1 (HSV-1) VP5, P-SSP7 gp10, T7 gp10, and λ gpE. (The HK97, T4, and HSV-1 images were adapted from reference 33 by permission from Macmillan Publishers Ltd.; the ϕ29 image was adapted from reference 60 with permission of the publisher; the ε15 image was adapted from reference 32 with permission from Macmillan Publishers Ltd.; the P22 image was adapted from reference 16 with permission of the publisher; the P-SSP7 image was adapted from reference 53 with permission of Macmillan Publishers Ltd.; the T7 image was adapted from reference 1 with permission of the publisher; and the λ image was adapted from reference 46 with permission of the publisher.)
Connector proteins.
The head-to-tail connecting region, termed the connector, ensures the cohesion of the phage capsid with its tail in all members of the Caudovirales and is often made of three different components organized as successive rings: the portal protein and two head completion proteins.
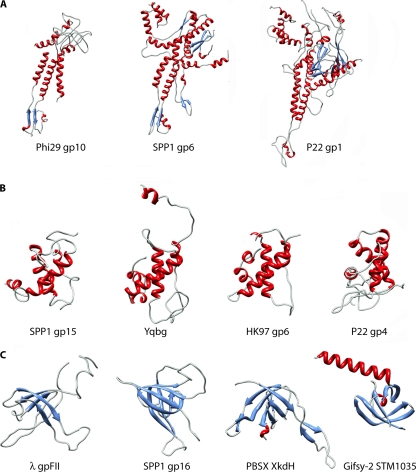
The portal is a keystone protein, located at one unique vertex of the viral capsid, involved in DNA packaging during assembly and allowing release at the onset of infection. Portal proteins from different phages do not have detectable sequence similarity and show large variations in their subunit molecular masses, from 37 kDa (for ϕ29) to 83 kDa (for P22). However, the plethora of structural data reported for portal proteins in all three Caudovirales families and also herpesviruses clearly demonstrated the conservation of the dodecameric core architecture of this phage component (which is shaped as a turbine) as well as of the folding pattern of the constituent monomers (19, 47, 48, 65, 76, 83) (Fig. 3A). In more detail, the lower portal moiety (termed the stalk) corresponds to the conserved scaffold, while the upper region (the wing) varies significantly in shape and size.
Fig. 3.
Conservation of the protein modules constituting bacteriophage connectors. (A) Gallery of portal protein structures determined by X-ray diffraction: ϕ29 gp10 (PDB accession no. 1FOU), SPP1 gp6 (PDB accession no. 2JES), and P22 gp1 (PDB accession no. 3LJ4). (B) Crystal structures of SPP1 gp15 (PDB accession no. 2KBZ), PBSX YqbG (PDB accession no. 1XN8), HK97 gp6 (PDB accession no. 3JVO), and P22 gp4 (PDB accession no. 3LJ4). (C) Crystal structures of λ gpFII (PDB accession no. 1K0H), SPP1 gp16 (PDB accession no. 2KCA), PBSX XkdH (PDB accession no. 3F3B), and Gifsy-2 STM1035 (PDB accession no. 2PP6). The coloring scheme used is based on secondary structures: blue, β-strands; red, α-helices; cyan, loops.
The middle ring of the phage connector has been reported to be constituted of very different proteins in terms of both sequence and structure, as illustrated by phage SPP1 gp15 and phage λ gpW (13, 52, 56). Nevertheless, the SPP1 gp15 fold is conserved in the siphophage HK97 gp6, in the podophage P22 gp4, as well as in YqbG, belonging to a PBSX-like prophage found in the Bacillus subtilis genome and forming Myoviridae particles (12, 66) (Fig. 3B).
Lastly, the bottom ring of the connector, acting as a plug maintaining DNA inside the capsid in SPP1, is assembled from a protein that is highly conserved among the Siphoviridae and Myoviridae families whose members of known structures are SPP1 gp16, PBSX XkdH, λ gpFII, and Gifsy-2 STM1035 (13, 52, 55) (Fig. 3C). A functional specialization has given rise to two distinct groups of this protein component based on the presence or absence of a long N-terminal extension correlated with the requirement to interact with a λ gpW-like or an SPP1 gp15-like partner, respectively (13).
Therefore, each of the three components encountered in many phage connectors (i.e., the portal and the two head completion proteins) appears to exhibit a distant respective ancestor, even though two evolutionarily distinct families seem to account for the proteins located in the middle ring of the connector.
Tail proteins.
The Siphoviridae tail architecture is rather simple and is based on three components: the central TMP, the tail tube protein or MTP, and the tail terminator protein. These components are also present and assembled in a similar way in Myoviridae tails, in addition to the sheath protein that provides the contractile nature to this organelle (2, 41, 42, 50).
Long-tailed bacteriophages require a tight regulation of the tail tube length, and this is achieved through the use of a ruler protein, i.e., the TMP. All Siphoviridae and Myoviridae genomes thus bear a large gene (>2 kbp) encoding such a protein that is characterized by the presence of extremely long hydrophobic α-helices flanked by two globular domains according to secondary-structure predictions. It has been shown that truncations or duplications of part of the TMP in phage λ, TP901-1, or TM4 are correlated with a reduced or increased tail tube length, respectively (38, 67, 71). Also, the overall physical length of the tail tubes of various phages is correlated directly with the length of the TMP-encoding gene (38). The physicochemical properties of TMPs make them a difficult target for structural studies, and no information has been reported up to now concerning this crucial component of bacteriophage tails.
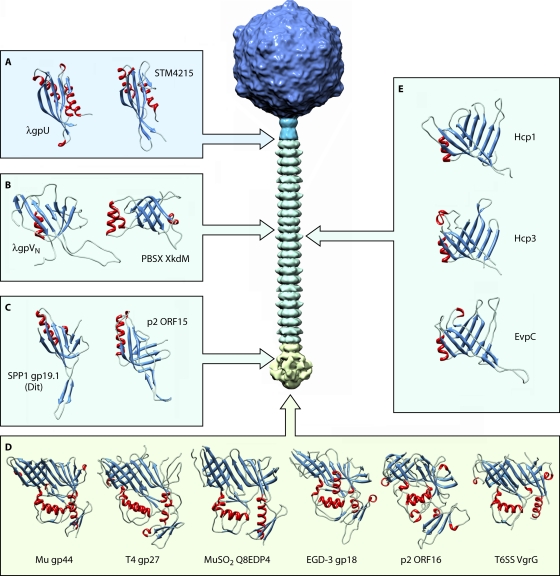
The second component of the phage tail tube is the tail terminator that acts in synergy with the TMP to stop tail tube polymerization when the genetically encoded length is reached by forming its most proximal ring. The tail terminator protein structures are known to be identical, both at tertiary and quaternary levels, between phage λ gpU and a putative protein (STM4215) found in the Salmonella enterica serovar Typhimurium genome (Fig. 4A) that is likely a typical representative of Myoviridae terminators (23, 70). This argues for the existence of a common ancestor of tail terminators encoded by long-tailed phages.
Fig. 4.
Conservation of the protein modules constituting bacteriophage tails and the T6SS apparatus needle. (A) Crystal structures of the tail terminator proteins λ gpU (PDB accession no. 3FZ2) and PBSX XkdM (PDB accession no. 2GJV). (B) Structures of the tail tube proteins of siphophage λ (gpVN [PDB accession no. 2K4Q]) and myophage PBSX (XkdM [PDB accession no. 2GUJ]) (C) Crystal structures of the Dit proteins SPP1 gp19.1 (PDB accession no. 2X8K) and p2 ORF15 (PDB accession no. 2WZP). (D) Gallery of gp27-like protein structures observed for the Myoviridae, including Mu gp44 (PDB accession no. 1WRU), T4 gp27 (PDB accession no. 1K28), and MuSO2 Q8EDP4 (PDB accession no. 3CDD); for the Siphoviridae, including EGD-e gp18 (PDB accession no. 3GS9) and p2 ORF16 (PDB accession no. 2WZP); as well as for the E. coli T6SS, including CFT073 VgrG (PDB accession no. 2P5Z). (E) Phage tail tube-like proteins from the T6SS tube: Hcp1 (PDB accession no. 1Y12) and Hcp3 (PDB accession no. 3HE1) from Pseudomonas aeruginosa and EVPC from Edwardsiella tarda (PDB accession no. 3EAA).
The monomeric structure of the phage λ MTP N-terminal domain (gpVN) combined with data from bioinformatic analyses has provided compelling evidence for a conserved architecture of this protein between sipho- and myophages (69). Indeed, the structural resemblance between the latter protein domain and XkdM, the PBSX tail tube protein, is remarkable, with both proteins having essentially the same topology and a presumed evolutionary relatedness (Fig. 4B). As MTP N-terminal domains are likely to be similarly folded in virtually all long-tailed phages and considering that this domain is responsible for the propagation of the firing signal upon binding to the host, it is possible that, in addition to being derived from a unique ancestral protein module, the domino-like signal transduction cascade mechanism proposed for SPP1 might apply to the Siphoviridae and the Myoviridae in general (3, 72). Various C-terminal domains are attached to this conserved module among phages: Ig-like domains, which are prominent (SPP1 gp17.1* FN3 [3] and λ gpV BIG-2 or I-set [28, 68]), RBP-like domains (Q54 [27]), or short orphan segments (PSA Tsh-L [92]). These different C-terminal domains cause the observed variation in the tail tube diameters of different phages and have been suggested to facilitate first contact and adhesion to the bacterial surface during the initial stages of infection. This diversity is likely the result of horizontal transfer events between phage structural proteins and bacterial modules (3, 27, 28).
The apparent common structure and evolution of contractile and noncontractile long tails of bacteriophages, as supported by the growing body of evidence presented here, raises intriguing questions as to whether the sheath protein evolved to interact with the tail tube of a common progenitor phage or whether a myophage-like progenitor lost its sheath to give rise to the family Siphoviridae.
The host adsorption apparatus.
At the distal tail end, a special device (varying in size, composition, and morphology) dedicated to specific host recognition is found, which can be as simple as a tail tip or in some cases consists of a larger macromolecular complex termed the baseplate. This organelle is the control center for infectivity. At first glance, looking at overall bacteriophage distal tail structure morphologies and architectures, it appears that large differences are present in this region, which seem to be correlated to the host adsorption strategy. Despite these major anatomical differences, a careful examination of baseplate and tail tip organizations reveals that common scaffolding principles also apply to these structural elements.
The conical structure originally identified in the baseplates of P335 species lactococcal phages, corresponding to the Dit protein, is the keystone of Gram-positive-bacterium-infecting Siphoviridae distal tail structures and plays an essential role in tail morphogenesis by priming initiation complex formation (57, 86, 87). It should be pointed out that no evidence for the presence of such a protein has been detected to date in siphophages targeting Gram-negative bacteria, whose tail tips hence appear to constitute a distinct class. Crystal structures of SPP1 Dit (85) and of p2 ORF15 (75) illustrate the remarkable conservation in this protein family, exhibiting virtually identical tertiary and quaternary structures, except for the “arm” extension observed for p2 (the RBP attachment sites) (Fig. 4C). Our results also indicate that an equivalent protein is present in the highly elaborated TP901-1 baseplate (7, 85). Considering that an identical Dit structure is present in phages as distant as SPP1 (bearing a simplified tail tip and interacting with a membrane protein receptor) and the P335 and 936 lactococcal phages (harboring large baseplates and adsorbing only to saccharidic components present in the host cell envelope) as well as the sequence similarity observed for this protein among several Gram-positive-bacterium-infecting phages (85), we postulate that a Dit-like protein with exactly the same architectural motif is found in all members of the Siphoviridae infecting Gram-positive bacteria. A genomic analysis of Dit proteins belonging to several phages infecting Gram-positive bacteria revealed that although the first half of the protein is highly conserved, some variability in terms of sequence and length are observed in the C-terminal moiety (85). We propose that this could reflect the adaptation of the protein to the various structural contexts of these phages, reflecting the need for variation in the adsorption apparatus while retaining the common topology of the N domain required to fit onto and interact with the tail tube.
Comparisons of gp27-like protein structures from Myoviridae phages T4 (gp27 [37]), Mu (gp44 [40]), and Shewanella oneidensis prophage MuSO2 (Q8EDP4 [Protein Data Bank {PDB} accession no. 3CDD]) as well as from Siphoviridae phage p2 (ORF16 [75]) and a Listeria monocytogenes prophage, EGD-e (gp18 [PDB accession no. 3GS9]), demonstrate that all these proteins are structurally similar and assemble as identical trimers (Fig. 4D). It is worth mentioning that a similar scaffold is thus present in totally different architectural schemes: gp27-like proteins are either directly exposed to the environment at the bottom of the baseplate (p2) or prolonged by the gp5 puncturing device (T4). The absence of a needle structure at the end of the phage tail in some bacteriophages raises questions about the mechanisms underlying membrane puncturing and/or cell wall digestion. In the case of P335-like phages TP901-1 and Tuc2009, and likely many other member of this group, a tail fiber with a hydrolytic activity has been identified, providing insights into the host perforation strategy (39, 86). Indeed, when these phages are committed to the host surface, the release of the Tal (tail-associated lysozyme) C-terminal enzymatic part allows the digestion of the bacterial cell wall and provides a pathway for DNA to cross the peptidoglycan during infection. The electron microscopy (EM) reconstruction of the TP901-1 baseplate that we recently reported suggests that the Tal (ORF47) N-terminal domain adopts the same fold as that of all other above-mentioned phages (7). Furthermore, PSI-BLAST analyses and transitive homology approaches (8) have demonstrated that the first 400 amino acid residues of several such proteins do share similar topologies (A. Davidson, personal communication). The combination of these data yields additional evidence that this module found in the Siphoviridae and Myoviridae has originated from a common ancestor.
Analysis of various Gram-positive phage antireceptor sequences reveals a similar general organization, suggesting a conserved strategy for phage diversification, in terms of host range, within several subspecies. An alignment of several Streptococcus thermophilus phage RBP sequences showed a highly conserved N-terminal moiety, probably due to its involvement in interactions with other tail proteins, whereas the C-terminal portion diverges in a region termed VR2 (variable region 2) that was proposed to recognize the host (20). Lactococcal phage RBPs also exhibit such characteristics, with a high degree of sequence conservation of both shoulder and neck domains in a given species (e.g., P335 or 936), while the head (receptor-binding) domain diverges in correlation with the different host ranges (21, 22, 57, 73). This pattern of sequence conservation, also observed for tail fibers of members of the Siphoviridae, Podoviridae, and Myoviridae infecting Gram-negative hosts, highlights that the part of the tail adsorption protein that contacts the host is under heavy evolutionary pressure to diversify as they are presented to a genetically changing target in the host cell population (6, 30). Hence, it appears that due to high selective pressure, tail fiber genes evolve more rapidly than other phage genes and that exchanges occur via horizontal transfer among phages crossing host phylogenetic boundaries (31).
Phage Evolution Strategies
As mentioned above, the “modular theory” of phage evolution states that evolution takes place through the transfer of exchangeable units that might consist only of protein domains rather than of whole genes or transcriptional units. Module exchange occurs at frequencies higher than or equal to the mutation rate, and it thus offers an enhanced genetic adaptation efficiency relative to evolution by linear descent (9). It is widely accepted that horizontal gene transfer events have occurred between large host phylogenetic distances and account for the high sequence similarities observed among genes present in phages targeting unrelated hosts (31). All dsDNA phage and prophage genomes seem therefore to be mosaic, with access by horizontal transfer to a large pool of genes, and the frequency of these events depends on the number of steps of genetic exchange required. The currently accumulated data validate this hypothesis, which is further illustrated by several cases such as (i) the conservation of a similar folding pattern in the head domain of lactococcal phage RBPs as well as in fiber proteins from reoviruses and adenoviruses, despite the absence of detectable sequence identity at the amino acid level (73, 78, 79, 84); (ii) the production of a chimeric RBP between the lactococcal phages TP901-1 and p2, in which no major structural changes occurred to accommodate the domain grafting (77); (iii) the observation of similar overall folds and organizations in the tailspikes of bacteriophages P22 (infecting Salmonella), HK620 (infecting Escherichia coli H), and Sf6 (infecting Shigella) that exhibit a high sequence conservation in the N-terminal virion-binding domain and no sequence identity in the receptor-binding domain (6, 51, 62, 80, 81); (iv) evidence of repeated tail fiber horizontal gene transfer among unrelated phages belonging to both the Siphoviridae and Myoviridae families (30); and (v) the widespread occurrence of three distinct Ig-like domain types (I-set, FN3, and BIG-2) in the three main Caudovirales families among phages infecting both Gram-positive and Gram-negative hosts (28).
In addition to the horizontal transfer of genetic material, phage adaptation is also likely to occur by point mutations in key genes (for example, those encoding tail fibers) in order to guarantee the recognition of hosts throughout their own evolution (84). Besides, protein processing from larger precursors is a strategy used by phages to determine phage-specific properties such as host range (54). Finally, the selective pressure exerted on phages by bacterial defense mechanisms, such as Abi (abortive-infection mechanism) (45) or CRISPR (clustered regularly interspaced short palindromic repeat) (29), may also be an important driving force of the genetic diversification of the bacteriophage population.
Same Fold, Different Functions
The wealth of structural, genomic, and functional data accumulated for the Siphoviridae and Myoviridae tail proteins have allowed us to infer specific properties relative to the evolutionary trajectory of these organelles. Remarkably, the tail tube protein (λ gpVN) fold was recently demonstrated to be similar to the Dit N-terminal domain one (85) and to the head-to-tail joining protein forming the most distal ring of the connector structure (e.g., λ gpFII, SPP1 gp16, PBSX XkdH, or Gifsy-2 STM1035) (12, 13, 55, 69, 75). These structures can also be superimposed with good agreement onto each of the two β-barrel domains of the gp27-like proteins (such as T4 gp27, Mu gp44, p2 ORF16, EGD-e gp18, and MuSO2 Q8EDP4) as well as onto a region of representative tail terminator structures (λ gpU and STM4215 present in the S. Typhimurium genome) (13, 37, 40, 75). Interestingly, the β-sheet that forms the inner wall of the continuous channel used for genome ejection into the host cell is strictly conserved among all these proteins. Another canonical feature of several of these proteins is the use of a loop acting as the belt extension, observed for the SPP1 Dit structure, to maintain the cohesion of these oligomeric assemblies. As the genes encoding all these components are located in close proximity to each other and in a conserved part of phage genomes, we extend the hypothesis formulated by Cardarelli et al. (12, 13) by proposing that these building blocks making the head completion protein found at the bottom of the connector, the tail terminator and tube, the Dit N-domain, as well as the gp27-like β-barrel domains might result from a unique ancestral protein by means of a series of duplication and diversification events. Indeed, despite a dramatic lack of sequence identity among many of their constituting proteins, all long phage tails appear to share an evolutionary origin. Our work combined with that of other groups (in particular A. Davidson's laboratory) provides an intriguing example of a very successful evolutionary mechanism for these proteins that do not have any sequence similarity and that perform different functions.
We also hypothesize that the high degree of sequence conservation observed at the level of the TMP C-terminal part, Dit, and the gp27-like N-terminal moiety among distantly related members of the Siphoviridae infecting Gram-positive hosts would result in similar structural motifs and conserved assembly mechanisms in such phages (85). Moreover, the combination of these observations with the structural data available for Dit-like and gp27-like family members lead us to propose that tail morphogenesis follows a similar pathway in the Siphoviridae and the Myoviridae (with some adaptations), with the formation of an initiation complex onto which other proteins attach (57, 70, 86). Finally, a likely hypothesis is that several of these components (e.g., λ gpV and gpU) as well as some head (e.g., λ gpD) and connector (e.g., λ gpW and gpFII as well as SPP1 gp15 and gp16) proteins adopt their quaternary structures only upon binding to their partners, with a regulatory scheme relying on the folding of unstructured regions inducing a disorder-to-order transition to allow a strict control of tail morphogenesis (12, 52, 55, 56, 69, 70). Therefore, the inability of these proteins to oligomerize on their own might provide a way to prevent aberrant assembly and nonproductive interactions.
BACTERIAL NEEDLELIKE STRUCTURES
Bacterial Secretion Systems
Bacterial secretion systems are large macromolecular assemblies releasing virulence factors into the external medium or translocating them directly into target cells. These virulence factors or effectors perform a large array of biochemical activities and modulate the function of crucial host regulatory molecules for the benefit of the producing bacterium. These systems are widespread in bacteria and provide them with the means to infect eukaryotic hosts and survive or replicate within them. Seven secretion systems have been documented, termed the type I secretion system (T1SS) to the T7SS, each involving 1 to 20 different proteins whose genes are often clustered together on so-called pathogenicity islands and transcribed as a single unit (14, 18). They often form a hollow conduit allowing the export of effectors (proteins and/or nucleic acids) through the complete cell envelope consisting of the cytoplasmic membrane, the peptidoglycan layer, and the outer membrane (where relevant) in a single step. These needlelike structures are evocative of bacteriophage tails in terms of morphology, size, and even function.
As for phage tail morphogenesis, a fundamental requirement of secretion systems is to control the length of the tube formed. It has been demonstrated that a ruler protein is responsible for size determination in the T3SS complex, a situation that is reminiscent of the mechanism of action of phage TMPs (35, 88, 89). Deletions or insertions performed in the central part of this protein result in injectisome shortening or lengthening, respectively, with a strict linear relationship observed between the needle length and the number of amino acids in the ruler (35). Both the N- and C-terminal parts of this protein are essential and have been proposed to serve as anchors. Therefore, the overall predicted architecture of the T3SS ruler appears to be identical to the typical bacteriophage TMP one. However, phage tails and T3SSs are distinguished by the fact that the former do not assemble in the absence of the ruler, whereas the latter do but with an undetermined length. It appears likely that this type of protein may be a requisite for the formation of various unrelated needlelike structures whose lengths must be regulated.
The Hcp protein family is an essential component of the T6SS, potentially forming the needle of this complex, and crystal structures of three such proteins have been reported: Hcp1 (PDB accession no. 1Y12) and Hcp3 (PDB accession no. 3HE1) from Pseudomonas aeruginosa as well as EVPC from Edwardsiella tarda (PDB accession no. 3EAA). These structures exhibit an identical fold, i.e., two antiparallel β-sheets plus an additional β-hairpin extension (Fig. 4E), all forming indistinguishable ∼90-Å-wide hexameric rings stabilized by a Dit-like belt extension and delineating a central channel with a 40-Å diameter (61). Moreover, the Hcp tertiary and quaternary structures are strikingly similar to Dit-like and MTP-like proteins and, thus, to the whole family of tail proteins described above (13, 69, 85). Notably, Hcp1 forms a tubular structure in its crystal lattice through a head-to-tail stacking of hexamers (61), and an Hcp homologue (Hcp3) was demonstrated to form tubes by EM (49). Hcp1 was also used to design tubes of various lengths by disulfide bridge engineering at the ring interfaces (5). All these structural data clearly establish an evolutionary connection between all these proteins sharing a similar fold and being involved in the formation of tube/needle assemblies with virtually identical dimensions (except tube length).
The crystal structure of the N-terminal portion of the T6SS protein VgrG (encompassing the first 483 out of 824 residues) from E. coli CFT073 reveals a topology virtually identical to that of T4 gp27 plus the gp5 OB fold domain, despite poor sequence conservation (49) (Fig. 4D). This structure, associated with bioinformatic results, suggests that in this secretion apparatus the two proteins forming the T4 puncturing device (gp27 and gp5) are fused in a unique polypeptide chain with the removal of the gp5 lysozyme domain (49). This difference is explained by the fact that VgrG is translocated into eukaryotic cells and therefore does not need a glycosidase activity to cross the cell envelope, in contrast to T4 gp5. The effector domains of many VgrG homologues are fused to their C termini, a situation that is reminiscent of the unexplained density observed for the T4 baseplate EM reconstruction at the level of the gp5 C-terminal β-helix tip (42, 49). In the case of Vibrio cholerae VgrG-1, the protein is thus a fusion of gp27, gp5, and the effector domain. By extension, the structural similarity also applies to p2 ORF16, Mu gp44, MuSO2 Q8EDP4, and EGD-e gp18, all possessing the T4 gp27-like topology (40, 75). In agreement with the observations described in the previous sections, the Hcp fold is reminiscent of the one observed for VgrG/gp27-like structures, and the Hcp hexameric ring can be superimposed onto the T4 gp27 trimeric pseudohexamer, creating a lineage between all those components (49).
Finally, a significant (∼40%) sequence similarity was detected between T4 gp25 and the E. coli CFT073 T6SS protein c3402 (49). As the former component contributes to the T4 baseplate structure by interacting with the (gp27-gp5)3 complex (at rest) or the tail tube (in the activated state), the presence of a homologous protein in the T6SS suggests that it plays a role in producing a baseplate-like assembly interacting with VgrG and Hcp.
The plethora of similarities observed between various components of bacteriophage tails and of bacterial secretion systems, in particular the T6SS, constitutes a converging set of clues toward the hypothesis of a common origin for all these devices that resemble each other at the structural and functional levels. Hence, pathogenic bacteria appear to have acquired the structural components of tailed bacteriophages in order to develop secretion systems that are crucial elements in support of their pathogenicity (36). Moreover, the accumulated data on bacteriophage tails and the T6SS support the idea that a single ancestral tail tube protein has yielded most of the proteins required to form a tube or a needle assembly by duplication events followed by specialization resulting from genetic exchange through insertions/deletions. It should be noted that assembly mechanisms similar to those observed for tail morphogenesis, involving disorder-to-order transitions within protein subunits, have also been proposed to contribute to T3SS formation (18) and, potentially, to T6SS formation (69).
Considering the observed structural and functional conservation, it is also conceivable that the domino-like signal transduction cascade, proposed for phage SPP1 and hypothetically applying to all long phage tails, is used in similar helical structures, such as the needle of bacterial secretion systems, in response to stimuli (72). Indeed, support for this notion was provided by work performed on T3SS, in which it was proposed that a contact with a target eukaryotic cell induces the transmission of a signal along the needle to activate the secretion machinery and results in effector translocation into the host cytoplasm (17, 18).
Bacteriocins
Beyond the resemblance observed between phage tails and secretion systems, other cases of bacteria that have evolved to use bacteriophage modules for their own advantage have been documented. A well-known example is that of bacteriocins, which are proteinaceous, antimicrobial compounds that are generally effective only against the same or closely related species. Two such cases will be discussed below: carotovoricins (Ctv) and pyocins.
Ctv are high-molecular-weight bacteriocins produced by several Erwinia carotovora strains and exhibiting a morphology outstandingly reminiscent of that of Myoviridae tails, with an antenna-like structure, a sheath, a core, a baseplate, and several tail fibers (64, 91). Genes encoding Ctv components are organized into clusters, and a survey of their amino acid sequences highlighted a high degree of similarity shared with various phage and prophage components (91). The tail fiber proteins found in these organelles exhibit the same modular organization as that observed for phages: the N-terminal part is highly conserved among Ctv fibers, probably to interact with the bacteriocin structure itself, whereas C-terminal extremities are highly divergent, thereby ensuring different killing spectra. Available data on Ctv led to the proposal that these bacteriocins were derived from a common myophage ancestor with some Salmonella prophages by losing head, lysogeny, and DNA replication modules. The diversity observed at the level of the tail fiber C-terminal end is thus probably a result of various horizontal transfer events among bacteriocins, phages, and prophages (91).
Pyocins are produced by Pseudomonas aeruginosa and can be divided into three types: R, F, and S (63). S-type pyocins are colicin-like proteins related to E2 colicin and will not be discussed here. R-type pyocins are reminiscent of myophage tails, and the various subtypes are differentiated by their receptor specificities, relying on the tail fiber proteins. F-type pyocins resemble siphophage tails and also feature several distinct receptor specificities among subtypes. Sequence comparisons showed extensive similarities between R-type pyocins and tails from phages belonging to the P2 group, whereas F-type pyocins are related to lambdoid phage tails (63). Furthermore, it was observed that within the clusters encoding these bacteriocins, the orders of the genes are extremely conserved relative to the corresponding phage tail-encoding regions. A lysis cassette, containing genes encoding an endolysin plus a holin as well as other accessory proteins (as found in bacteriophages), is also present within the bacteriocin-encoding regions, probably to allow the release of these bioactive molecules. These observations point to an evolutionary connection between the R-type or F-type pyocins and P2-like or λ-like bacteriophages, respectively. It was thus postulated that both types of pyocins represent bacteriocins that are evolutionarily specialized rather than defective phages (63). The tail fiber organization found in pyocins follows the general principle described for phages and Ctv: a conserved N-terminal part is responsible for interactions with the needlelike structure, while a hypervariable C-terminal region mediates specific receptor recognition, allowing different pyocins to target different hosts. Remarkably, regulatory systems of pyocin expression are redolent of temperate bacteriophages: UV irradiation or mitomycin treatment induces the expression of pyocin genes (63).
CONCLUSIONS
The availability of a large number of bacteriophage protein structures has made it possible to analyze the evolutionary relationships existing among virions belonging to the order Caudovirales and even to other more distant viruses. The data presented here strongly suggest that a single protein module has given rise to most of the proteins forming the various phage tail components as well as other needlelike assemblies, such as secretion systems and bacteriocins. Together, these observations emphasize the importance of phage-derived elements in the evolution and function of diverse and complex bacterial systems and in bacterial adaptation to new environmental conditions. Indeed, the acquisition and specialization of bacteriophage genetic modules are important means used by bacteria to develop new functions that ultimately provide a distinct selective advantage.
ACKNOWLEDGMENTS
This work was supported, in part, by grants from the Marseille-Nice Génopole, the CNRS, and the ANR (ANR-07-BLAN-0095) and by a Ph.D. grant from the Ministère Français de l'Enseignement Supérieur et de la Recherche to D.V. (reference no. 22976-2006).
We are very grateful to Alan Davidson and Sylvain Moineau for fruitful discussions.
Biographies
David Veesler obtained a master's and a Ph.D. in Structural Biology at the Architecture et Fonction des Macromolécules Biologiques laboratory in Marseille, France (Université de Provence). During this period his work was dedicated to the study of the initial events underlying bacteriophage infection using a combination of structural and biophysical methods. He is currently working as a Research Associate at The Scripps Research Institute and focuses on bacteriophage capsid maturation. His main interests are the structural studies of large macromolecular complexes using hybrid methods (X-ray crystallography, electron microscopy, and light scattering, etc.).
Christian Cambillau obtained a Ph.D. in Chemistry at the University of Orsay (Paris-South) in 1978. After a postdoctoral stay in Uppsala, Sweden, with Prof. C.-I. Brändèn, he joined a protein crystallography group in Marseille, France. During this period, his work was dedicated to molecular graphics and crystallographic studies of lectins, lipases, redox, olfactory proteins, and camelid antibodies. He was head of the Architecture et Fonction des Macromolécules Biologiques Structural Biology laboratory between 1990 and 2004. He is currently working in this laboratory as a group leader, where, after being involved in large Structural Genomics projects on viral enzymes, he turned to the structural study of large complexes from phages (>1 MDa) and secretion systems using hybrid methods (X-ray crystallography, electron microscopy, and light scattering).
REFERENCES
- 1. Agirrezabala X., et al. 2007. Quasi-atomic model of bacteriophage t7 procapsid shell: insights into the structure and evolution of a basic fold. Structure 15:461–472 [DOI] [PubMed] [Google Scholar]
- 2. Aksyuk A. A., et al. 2009. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 28:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Auzat I., Droge A., Weise F., Lurz R., Tavares P. 2008. Origin and function of the two major tail proteins of bacteriophage SPP1. Mol. Microbiol. 70:557–569 [DOI] [PubMed] [Google Scholar]
- 4. Baker M. L., Jiang W., Rixon F. J., Chiu W. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79:14967–14970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ballister E. R., Lai A. H., Zuckermann R. N., Cheng Y., Mougous J. D. 2008. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U. S. A. 105:3733–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barbirz S., et al. 2008. Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol. Microbiol. 69:303–316 [DOI] [PubMed] [Google Scholar]
- 7. Bebeacua C., et al. 2010. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J. Biol. Chem. 285:39079–39086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bolten E., Schliep A., Schneckener S., Schomburg D., Schrader R. 2001. Clustering protein sequences—structure prediction by transitive homology. Bioinformatics 17:935–941 [DOI] [PubMed] [Google Scholar]
- 9. Botstein D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484–490 [DOI] [PubMed] [Google Scholar]
- 10. Bray J. E., Todd A. E., Pearl F. M., Thornton J. M., Orengo C. A. 2000. The CATH Dictionary of Homologous Superfamilies (DHS): a consensus approach for identifying distant structural homologues. Protein Eng. 13:153–165 [DOI] [PubMed] [Google Scholar]
- 11. Brussow H., Hendrix R. W. 2002. Phage genomics: small is beautiful. Cell 108:13–16 [DOI] [PubMed] [Google Scholar]
- 12. Cardarelli L., et al. 2010. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J. Mol. Biol. 395:754–768 [DOI] [PubMed] [Google Scholar]
- 13. Cardarelli L., et al. 2010. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc. Natl. Acad. Sci. U. S. A. 107:14384–14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cascales E. 2008. The type VI secretion toolkit. EMBO Rep. 9:735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chandry P. S., Moore S. C., Boyce J. D., Davidson B. E., Hillier A. J. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49–64 [DOI] [PubMed] [Google Scholar]
- 16. Chen D. H., et al. 2011. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. U. S. A. 108:1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cordes F. S., et al. 2003. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 278:17103–17107 [DOI] [PubMed] [Google Scholar]
- 18. Deane J. E., et al. 2006. Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc. Natl. Acad. Sci. U. S. A. 103:12529–12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doan D. N., Dokland T. 2007. The gpQ portal protein of bacteriophage P2 forms dodecameric connectors in crystals. J. Struct. Biol. 157:432–436 [DOI] [PubMed] [Google Scholar]
- 20. Duplessis M., Moineau S. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325–336 [DOI] [PubMed] [Google Scholar]
- 21. Dupont K., Janzen T., Vogensen F. K., Josephsen J., Stuer-Lauridsen B. 2004. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl. Environ. Microbiol. 70:5825–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dupont K., Vogensen F. K., Neve H., Bresciani J., Josephsen J. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ. Microbiol. 70:5818–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edmonds L., et al. 2007. The NMR structure of the gpU tail-terminator protein from bacteriophage lambda: identification of sites contributing to Mg(II)-mediated oligomerization and biological function. J. Mol. Biol. 365:175–186 [DOI] [PubMed] [Google Scholar]
- 24. Effantin G., Boulanger P., Neumann E., Letellier L., Conway J. F. 2006. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J. Mol. Biol. 361:993–1002 [DOI] [PubMed] [Google Scholar]
- 25. Effantin G., Figueroa-Bossi N., Schoehn G., Bossi L., Conway J. F. 2010. The tripartite capsid gene of Salmonella phage Gifsy-2 yields a capsid assembly pathway engaging features from HK97 and lambda. Virology 402:355–365 [DOI] [PubMed] [Google Scholar]
- 26. Fokine A., et al. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U. S. A. 102:7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fortier L. C., Bransi A., Moineau S. 2006. Genome sequence and global gene expression of Q54, a new phage species linking the 936 and c2 phage species of Lactococcus lactis. J. Bacteriol. 188:6101–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fraser J. S., Yu Z., Maxwell K. L., Davidson A. R. 2006. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J. Mol. Biol. 359:496–507 [DOI] [PubMed] [Google Scholar]
- 29. Garneau J. E., et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71 [DOI] [PubMed] [Google Scholar]
- 30. Haggard-Ljungquist E., Halling C., Calendar R. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hendrix R. W., Smith M. C., Burns R. N., Ford M. E., Hatfull G. F. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang W., et al. 2008. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature 451:1130–1134 [DOI] [PubMed] [Google Scholar]
- 33. Jiang W., et al. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang W., et al. 2003. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 10:131–135 [DOI] [PubMed] [Google Scholar]
- 35. Journet L., Agrain C., Broz P., Cornelis G. R. 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302:1757–1760 [DOI] [PubMed] [Google Scholar]
- 36. Kanamaru S. 2009. Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc. Natl. Acad. Sci. U. S. A. 106:4067–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kanamaru S., et al. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553–557 [DOI] [PubMed] [Google Scholar]
- 38. Katsura I., Hendrix R. W. 1984. Length determination in bacteriophage lambda tails. Cell 39:691–698 [DOI] [PubMed] [Google Scholar]
- 39. Kenny J. G., McGrath S., Fitzgerald G. F., van Sinderen D. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J. Bacteriol. 186:3480–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kondou Y., et al. 2005. Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J. Mol. Biol. 352:976–985 [DOI] [PubMed] [Google Scholar]
- 41. Kostyuchenko V. A., et al. 2005. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 12:810–813 [DOI] [PubMed] [Google Scholar]
- 42. Kostyuchenko V. A., et al. 2003. Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Biol. 10:688–693 [DOI] [PubMed] [Google Scholar]
- 43. Krupovic M., Bamford D. H. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 6:941–948 [DOI] [PubMed] [Google Scholar]
- 44. Krupovic M., Forterre P., Bamford D. H. 2010. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 397:144–160 [DOI] [PubMed] [Google Scholar]
- 45. Labrie S. J., Samson J. E., Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 46. Lander G. C., et al. 2008. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure 16:1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lander G. C., et al. 2009. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure 17:789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lebedev A. A., et al. 2007. Structural framework for DNA translocation via the viral portal protein. EMBO J. 26:1984–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leiman P. G., et al. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leiman P. G., Chipman P. R., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmann M. G. 2004. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118:419–429 [DOI] [PubMed] [Google Scholar]
- 51. Leiman P. G., Molineux I. J. 2008. Evolution of a new enzyme activity from the same motif fold. Mol. Microbiol. 69:287–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lhuillier S., et al. 2009. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc. Natl. Acad. Sci. U. S. A. 106:8507–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu X., et al. 2010. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat. Struct. Mol. Biol. 17:830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lubbers M. W., Waterfield N. R., Beresford T. P., Le Page R. W., Jarvis A. W. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4348–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maxwell K. L., Yee A. A., Arrowsmith C. H., Gold M., Davidson A. R. 2002. The solution structure of the bacteriophage lambda head-tail joining protein, gpFII. J. Mol. Biol. 318:1395–1404 [DOI] [PubMed] [Google Scholar]
- 56. Maxwell K. L., et al. 2001. The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold. J. Mol. Biol. 308:9–14 [DOI] [PubMed] [Google Scholar]
- 57. Mc Grath S., et al. 2006. Anatomy of a lactococcal phage tail. J. Bacteriol. 188:3972–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Moineau S., Pandian S., Klaenhammer T. R. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mondigler M., Ayoub A. T., Heller K. J. 2006. The DNA region of phage BF23 encoding receptor binding protein and receptor blocking lipoprotein lacks homology to the corresponding region of closely related phage T5. J. Basic Microbiol. 46:116–125 [DOI] [PubMed] [Google Scholar]
- 60. Morais M. C., et al. 2005. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol. Cell 18:149–159 [DOI] [PubMed] [Google Scholar]
- 61. Mougous J. D., et al. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Muller J. J., et al. 2008. An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri phage Sf6. Structure 16:766–775 [DOI] [PubMed] [Google Scholar]
- 63. Nakayama K., et al. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213–231 [DOI] [PubMed] [Google Scholar]
- 64. Nguyen A. H., Tomita T., Hirota M., Sato T., Kamio Y. 1999. A simple purification method and morphology and component analyses for carotovoricin Er, a phage-tail-like bacteriocin from the plant pathogen Erwinia carotovora Er. Biosci. Biotechnol. Biochem. 63:1360–1369 [DOI] [PubMed] [Google Scholar]
- 65. Olia A. S., Casjens S., Cingolani G. 2007. Structure of phage P22 cell envelope-penetrating needle. Nat. Struct. Mol. Biol. 14:1221–1226 [DOI] [PubMed] [Google Scholar]
- 66. Olia A. S., Prevelige P. E., Jr., Johnson J. E., Cingolani G. 2011. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 18:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pedersen M., Ostergaard S., Bresciani J., Vogensen F. K. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315–328 [DOI] [PubMed] [Google Scholar]
- 68. Pell L. G., et al. The solution structure of the C-terminal Ig-like domain of the bacteriophage lambda tail tube protein. J. Mol. Biol. 403:468–479 [DOI] [PubMed] [Google Scholar]
- 69. Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U. S. A. 106:4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pell L. G., et al. 2009. The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J. Mol. Biol. 389:938–951 [DOI] [PubMed] [Google Scholar]
- 71. Piuri M., Hatfull G. F. 2006. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 62:1569–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Plisson C., et al. 2007. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 26:3720–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ricagno S., et al. 2006. Crystal structure of the receptor-binding protein head domain from Lactococcus lactis phage bIL170. J. Virol. 80:9331–9335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rohwer F., Edwards R. 2002. The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sciara G., et al. 2010. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. U. S. A. 107:6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Simpson A. A., et al. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Siponen M., et al. 2009. Crystal structure of a chimeric receptor binding protein constructed from two lactococcal phages. J. Bacteriol. 191:3220–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Spinelli S., et al. 2006. Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1. J. Biol. Chem. 281:14256–14262 [DOI] [PubMed] [Google Scholar]
- 79. Spinelli S., et al. 2006. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat. Struct. Mol. Biol. 13:85–89 [DOI] [PubMed] [Google Scholar]
- 80. Steinbacher S., et al. 1997. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267:865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Steinbacher S., et al. 1994. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265:383–386 [DOI] [PubMed] [Google Scholar]
- 82. Sutter M., et al. 2008. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 15:939–947 [DOI] [PubMed] [Google Scholar]
- 83. Tang L., Marion W. R., Cingolani G., Prevelige P. E., Johnson J. E. 2005. Three-dimensional structure of the bacteriophage P22 tail machine. EMBO J. 24:2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tremblay D. M., et al. 2006. Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 188:2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Veesler D., et al. 2010. Crystal structure of bacteriophage SPP1 distal tail protein (gp19.1): a baseplate hub paradigm in Gram-positive infecting phages. J. Biol. Chem. 285:36666–36673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vegge C. S., et al. 2005. Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 187:4187–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vegge C. S., et al. 2006. Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009. J. Bacteriol. 188:55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wagner S., et al. 2009. The helical content of the YscP molecular ruler determines the length of the Yersinia injectisome. Mol. Microbiol. 71:692–701 [DOI] [PubMed] [Google Scholar]
- 89. Wagner S., Stenta M., Metzger L. C., Dal Peraro M., Cornelis G. R. 2010. Length control of the injectisome needle requires only one molecule of Yop secretion protein P (YscP). Proc. Natl. Acad. Sci. U. S. A. 107:13860–13865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Xu J., Hendrix R. W., Duda R. L. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11–21 [DOI] [PubMed] [Google Scholar]
- 91. Yamada K., et al. 2006. Nucleotide sequences and organization of the genes for carotovoricin (Ctv) from Erwinia carotovora indicate that Ctv evolved from the same ancestor as Salmonella typhi prophage. Biosci. Biotechnol. Biochem. 70:2236–2247 [DOI] [PubMed] [Google Scholar]
- 92. Zimmer M., Sattelberger E., Inman R. B., Calendar R., Loessner M. J. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed +1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50:303–317 [DOI] [PubMed] [Google Scholar]