Abstract
Porcine circovirus type 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) are major contributors to the porcine respiratory disease complex (PRDC). Routine serological diagnosis and surveillance play an important role in the prevention of PRDC, as it is a leading cause of economic losses to the swine industry. We herein describe an advanced microsphere-based immunoassay that permits the simultaneous detection of antibodies to PCV2 and PRRSV, thereby reducing the time and effort involved in testing. Recombinant PRRSV nucleoprotein antigen and the PCV2 capsid antigen were coupled to fluorophore-dyed beads with distinct spectral addresses. Weekly serum samples from 72 pigs that were experimentally exposed to either PCV2, PRRSV, or both PCV2 and PRRSV were used to validate the microbead assay (MBA) in comparison with the “gold standard” enzyme-linked immunosorbent assays. The kinetics of the PCV2- and PRRSV-specific antibody responses measured by the microbead assay were comparable to those of the standard assays; Spearman's rank correlations were 0.72 (P < 0.001) for PRRSV and 0.80 (P < 0.001) for PCV2. Diagnostic sensitivity and specificity were determined using field sera whose positive or negative status was determined by the standard tests. The diagnostic sensitivity and specificity were both 98% for PCV2 and were 91% and 93%, respectively, for PRRSV (kappa coefficients, 0.85 and 0.67 for PCV2 and PRRSV, respectively). Multiplexing did not interfere with assay performance or diagnostic sensitivity. Therefore, the described study demonstrates proof of concept for the development of more versatile and economical microbead array-based multiplex serological test panels for veterinary use.
INTRODUCTION
Porcine respiratory disease complex (PRDC) is a multifactorial disease syndrome which can involve several etiological agents. Therefore, early detection and prevention of coinfections are important aspects of managing PRDC. Growing piglets are the most severely affected, with PRDC resulting in morbidity which can be as high as 70%, poor feed conversion, and lower growth rates. Porcine circovirus type 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) are the most frequently associated with PRDC. Swine influenza viruses (SIVs), porcine respiratory coronavirus, Mycoplasma spp., Actinobacillus pleuropneumoniae, Streptococcus suis, and Pasteurella spp. may also be etiological agents (3, 14, 20, 36).
Due to the severe economic losses associated with PRDC, serological testing for agents involved in PRDC comprises a major part of the diagnostic testing carried out in swine-dense regions. In addition to the fact that multiple agents can be simultaneously involved in causing PRDC, agents like SIV and A. pleuropneumoniae have several subtypes or serotypes which are antigenically distinct (24, 37). In theory, a comprehensive serological test panel for the differential diagnosis of PRDC should incorporate tests for several different pathogens. The tests of choice for each of these agents could potentially include several different assay formats, such as the enzyme-linked immunosorbent assay (ELISA) and the more laborious complement fixation tests, hemagglutination inhibition, and virus neutralization tests (18). Therefore, multiplex testing platforms which can save labor, time, and cost by providing information about more than one pathogen from a single test run will be important as diagnostic tools of the future. While several multiplex tests have been developed for nucleic acid-based detection of pathogens (15, 29, 38), very few multiplex tests are available for antibody detection, especially for veterinary use (1, 8, 21).
The microbead array (MBA)-based technology (Luminex Corp., Austin, TX) consists of color-coded microbeads which have distinct spectral addresses enabling laser-mediated detection in a flow cytometer. The beads can be coupled to antigens, antibodies, or nucleic acids for the specific detection of cognate antibodies, antigens (such as cytokines), or DNA for diagnostic and basic research applications (28). Up to 100 different analytes can be detected simultaneously with the Luminex xMap technology. Therefore, the primary advantages of this technology are its multiplexing and high-throughput capabilities. Other advantages include savings of cost, time, and labor and improved assay performance. Interest in the MBA technology as a versatile tool for clinical microbiology with both human and animal applications is widespread and increasing (1, 13, 21, 23, 34). In this study, we have harnessed the MBA technology to develop a dual serological detection test for PCV2 and PRRSV (PCV2/PRRSV MBA), as a first step toward the development of a comprehensive multiplex test panel for PRDC.
MATERIALS AND METHODS
PCV2 and PRRSV antigens.
The immunogenic PCV2 capsid protein which is encoded by open reading frame 2 (orf2) was expressed in the baculovirus system as previously described (27). Nonrecombinant baculovirus antigen was used as a background control and is designated the wild-type (WT) antigen in this study. PRRSV nucleoprotein (N protein) was produced in a bacterial expression system as a 6×His-tagged protein and was purified by nickel affinity chromatography (19).
PCV2 and PRRSV standard ELISAs for validation of PCV2/PRRSV MBA.
The optimization and validation of the orf2-based PCV2 ELISA have been previously described (27). A slightly modified version is currently offered at the Iowa State University Veterinary Diagnostic Laboratory (ISU-VDL). Briefly, 16 ng of PCV2 Orf2 and 5.2 ng of WT antigen/well are coated on adjacent wells of an Immunolon plate (Dynex, Chantilly, VA). A 1:100 dilution of test serum in blocking buffer consisting of 5% skim milk powder is incubated on the plates for 30 min, followed by the addition of anti-swine horseradish peroxidase conjugate at 1:2,500. After incubation with the substrate and stopping of the reaction with 0.1 N H2SO4, plates are read at 450 nm. The corrected optical density (COD) value of each sample is derived by subtraction of the WT value from the orf2 value for the given sample. The COD value of each sample is divided by the COD value of the positive-control serum sample and expressed as a sample COD-to-positive-control COD (S/P) ratio. A cutoff value of 0.3 was used to distinguish between positive and negative samples. For the purposes of this study, samples with values between 0.2 and 0.3 were classified as suspect. As described by Nawagitgul et al. (27), the assay had a sensitivity of 91% and a specificity of 93%.
A well-accepted commercial ELISA kit (Herdchek PRRSV 2XR kit; IDEXX, Westbrook, ME) was used for the detection of PRRSV antibodies following the manufacturer's instructions. A manufacturer prescribed S/P cutoff value of 0.4 was used to distinguish between positive and negative samples. According to a previous study, the sensitivity and specificity values for the assay were determined to be 97.4% and 99.6%, respectively (4). However, users of the 2XR kit have consistently recognized and documented unexpected positive results (4, 7, 17).
Serum samples for assay validation.
Archived experimental serum samples from a previous study (33) were used to validate the PCV2/PRRSV MBA. Seventy-two PCV2-, PRRSV-, and SIV-free pigs were randomly assigned to four groups. Group 1 piglets (n = 12) served as uninfected controls; group 2 piglets (n = 24) were divided into two subgroups. Each pig in one subgroup of 12 piglets was inoculated with 104.5 50% tissue culture infective doses (TCID50) of the PCV2 wild-type virus strain 40895 (6) stock (3 ml intranasally and 2 ml intramuscularly), while the 12 pigs in the other subgroup were administered a mutant virus which had an altered interferon-stimulated response element (ISRE) in its genome (32). We have previously shown that the mutant virus and wild-type PCV2 are antigenically indistinguishable by the “gold standard” PCV2 ELISA (33). Therefore, no distinction is made between the two viruses for the purpose of validation of the PCV2/PRRSV MBA in this study. Data from animals infected with either the wild-type virus or the mutant PCV2 have been combined for analysis. Group 3 piglets (n = 12) were inoculated intranasally with 106.0 TCID50 of PRRSV strain VR2385 stock; group 4 pigs (n = 24) were coinfected with both PCV2 and PRRSV as described for groups 2 and 3. At 14 days postinfection (dpi), 6 pigs from groups 1 and 3 and 12 pigs each from the remaining groups were killed. All pigs were euthanized at 28 dpi. Sera which were collected just prior to infection at 1 dpi and weekly thereafter were used to validate and optimize the PCV2/PRRSV MBA.
The diagnostic sensitivity and specificity of the PCV2/PRRSV MBA were assessed using field samples (11, 12). Three hundred eighty-eight field samples were analyzed, and 226 were classified PCV2 negative (S/P > 0.2), 28 suspect (S/P = 0.2 to 0.3), and 134 positive (S/P > 0.3) on the basis of the in-house gold standard PCV2 ELISA. The selected negative samples had S/P values that ranged from −0.108 to 0.204, and the positive samples had S/P values that ranged from 0.296 to 3.13.
Three hundred seventy-eight samples were analyzed by the commercial HerdChek PRRS 2XR ELISA. Of these, 262 were PRRSV negative with S/P values that ranged from 0.00 to 0.399 and 116 samples were positive with S/P values that ranged from 0.404 to 4.26. There was no suspect range assigned to these samples, as it was not prescribed by the ELISA kit manufacturer. There were 124 samples that were assessed on both the PCV2 and PRRSV standard ELISAs (see Fig. 3). Selected samples from a previously published study were used to determine the cross-reactivity of the PCV2 and PRRSV antigens to SIV (22) (Table 1).
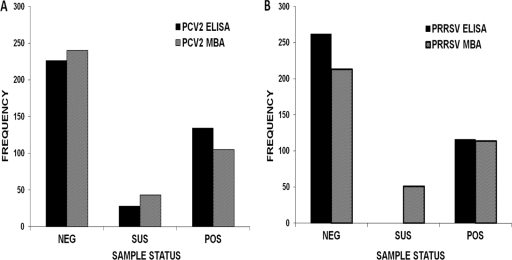
Fig. 3.
Distribution of the number of positive (POS), suspect (SUS), and negative (NEG) field samples, as classified by the gold standard ELISAs and the MBA at the cutoffs selected by ROC analysis. (A) Comparison of PCV2 ELISA and MBA; (B) comparison of PRRSV ELISA and MBA.
Table 1.
Results for control samples used to measure interassay variation of PCV2/PRRSV MBA
| Sample | Result fora: |
||
|---|---|---|---|
| PCV2 | PRRSV | SIV | |
| Control 1 | P | N | N |
| Control 2 | N | N | N |
| Control 3 (pooled controls 1 and 2) | P | N | N |
| Control 4 (PCV2 and PRRSV positive) | P | P | N |
| Control 5 (PCV2 positive) | P | N | N |
| Control 6 (SIV, PCV2, and PRRSV positive) | P | P | P |
| Control 7 (pooled controls 4, 5, and 6) | P | P | P |
P, positive; N, negative.
PCV2/PRRSV duplex microbead assay.
Twelve micrograms of the PCV2 Orf2 antigen, 8 μg of the PRRSV N-protein antigen, and 6 μg of the WT antigen were each coupled to fluorophore-dyed microbead numbers 65, 61, and 48 (Bio-Rad, Hercules, CA) as described by the manufacturer. Bead counts were verified on a hemocytometer. A total of 2,500 beads/well/antigen were used for all assays. The microbead assay was carried out as described previously, with modifications (1, 8, 34). The 96-well filter plates (Millipore, Billerica, MA) were blocked with PBN buffer (phosphate-buffered saline [PBS], pH 7.4, with 1% bovine serum albumin) for 2 min. Test sera were diluted 1:100 in PBN buffer and added to the filter plate. A total of 2,500 coupled beads for each of the three antigens used in the study were added to the wells. The plate was incubated at room temperature for 30 min, followed by three washes with PBS-Tween (PBS-T) using a vacuum manifold. Biotin-labeled anti-swine IgG H&L chain (Kirkegaard & Perry Laboratories, San Diego, CA) was added at 1:2,500, and the plate was incubated at room temperature for 30 min. Plates were washed three times with PBS-T. Fifty microliters of streptavidin conjugated with Rhodophyta phycoerythrin (Invitrogen, Valencia, CA) at a 1:100 dilution in PBN buffer was added for 30 min. The plate was again washed twice in a vacuum manifold. The beads were resuspended in 125 μl of PBN buffer. Seventy-five microliters of the resuspended beads was transferred to a clear U-bottom plate (Thermo Scientific, Worcester, MA). Plates were read in the Luminex 100 instrument using Bioplex (version 5.0) software (Bio-Rad, Hercules, CA). The median fluorescence intensity unit (MFU) for each of the three antigen-coupled beads in the wells was the median value for a count of 100 beads detected by the dual-flow laser cytometer (1, 8, 34). PCV2/PRRSV MBA data for the experimental samples used in this study are presented as the average of the MFU values for each treatment group. Therefore, they are denoted the average fluorescence intensity (AFU) value (see Fig. 1 and Table 2). A panel of seven selected serum samples was used as a control in all assays (Table 1).
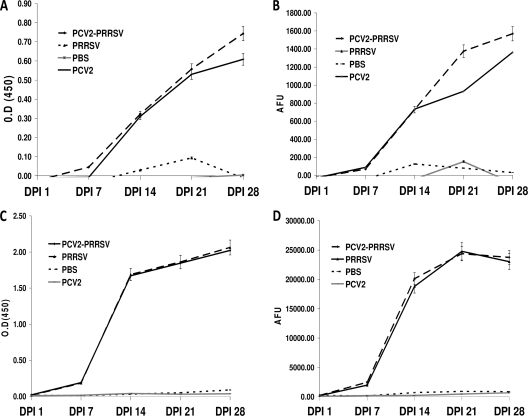
Fig. 1.
Kinetics of antibody responses in pigs that were singly or dually infected with PCV2 and/or PRRSV. (A) Antibody responses measured by the gold standard PCV2 ELISA; (B) antibody responses measured by the PCV2 MBA; (C) antibody responses measured by the gold standard PRRSV ELISA; (D) antibody responses measured by the PRRSV MBA.
Table 2.
Interassay variation of PCV2/PRRSV MBA
| Sample | AFU |
|||||
|---|---|---|---|---|---|---|
| PCV2 Orf2 WT |
PRRSV N protein |
|||||
| Mean | Median | COV | Mean | Median | COV | |
| Control 1 | 1,370.67 | 1,304.00 | 0.20 | 547.67 | 488.50 | 0.29 |
| Control 2 | −13.06 | −19.00 | −3.27 | 74.72 | 3.00 | 2.25 |
| Control 3 (pooled controls 1 and 2) | 1,053.39 | 1,041.00 | 0.26 | 373.61 | 354.00 | 0.35 |
| Control 4 (PCV2 and PRRSV positive) | 1,593.83 | 1,456.00 | 0.21 | 22,393.00 | 22,595.00 | 0.07 |
| Control 5 (PCV2 positive) | 1,686.67 | 1,641.50 | 0.28 | 21.11 | 26.00 | 2.28 |
| Control 6 (SIV, PCV2, and PRRSV positive) | 4,836.67 | 4,980.00 | 0.30 | 21,789.83 | 24,020.50 | 0.17 |
| Control 7 (pooled controls 4, 5, and 6) | 3,228.50 | 3,484.50 | 0.17 | 22,154.28 | 23,267.50 | 0.10 |
Optimization of PCV2/PRRSV MBA.
Three spectrally distinct microbead populations were selected for conjugation of PRRSV N protein, PCV2 Orf2, and WT protein at each of three amounts: 6 μg, 8 μg, and 12 μg. Three strongly positive and three negative experimental serum samples from 28 dpi in the study described previously (20) were pooled. Two SIV-positive and -negative samples each were included to control for cross-reactivity, as SIV is another common causative agent of PRDC. A partial checkerboard titration with 1:50, 1:100, and 1:200 dilutions of the primary antibodies and 1:500, 1:1,000, 1:1,500 1:2,000, and 1:2,500 dilutions of the biotin-labeled anti-swine IgG conjugate was assessed by the PCV2/PRRSV MBA.
Interassay variation was determined using the panel of seven control serum samples (Tables 1 and 2) which were positive for either PCV2 or PRRSV, or both, as previously described (24). One sample that was also positive for SIV was used to rule out cross-reactivity to SIV. Two of the seven controls were created by pooling to demonstrate that detection was not affected by either dilution of the positives or the presence of antibodies to more than one analyte. The samples were assessed in duplicate over eight different runs. Analytical specificity was evaluated as cross-reactivity between PCV2, PRRSV, and SIV. Intra-assay variation was measured by testing selected negative, low-positive, and high-positive sera in duplicate in different locations on the same plate. The coefficient of variation (COV) between readings for each sample was calculated as a measure of assay variation (11, 12).
Statistical analysis.
For the experimental samples, the correlation between the standard ELISAs and the PCV2/PRRSV MBA was assessed by using Spearman's correlation coefficient. To determine whether multiplexing interfered with detection of either analyte, the effect of the presence of PRRSV antibodies on MBA detection of PCV2 antibodies and vice versa was assessed by the Wilcoxon two-sample rank-sum test. For field samples, receiver operating characteristics (ROC) analysis (9–11) was carried out for each analyte using the ELISA as the standard test to determine the positive and negative status of a sample. Sensitivity- and specificity-controlled positive, negative, and suspect cutoff values were selected on the basis of the ROC analysis. Positive predictive values, negative predictive values, sensitivities, and specificities were calculated at the selected cutoff values. Agreement between dichotomized ELISA and dichotomized PCV2/PRRSV MBA data was assessed using the proportion of agreement and the kappa coefficient association.
RESULTS
Optimization of PCV2/PRRSV MBA.
On the basis of the results of the partial checkerboard titration, 12 μg of Orf2 antigen, 6 μg of WT antigen, and 8 μg of PRRSV N-protein antigen were determined to be optimal for bead coupling. A 1:100 dilution of the test serum provided an adequate dynamic range with optimal sensitivity for the detection of low-positive samples. The working conjugate dilution was set at 1:2,500. The lack of cross-reactivity of the PCV2 antigen to porcine parvovirus and PRRSV antibodies has been previously described (27). With a view to expanding multiplex testing capabilities using the PCV2/PRRSV MBA, we also examined the cross-reactivities of the selected antigens to anti-SIV antibodies. The analytical specificity of the assay was satisfactory, as no cross-reactivity between PCV2-, PRRSV-, and SIV-positive sera was detected on the PCV2/PRRSV MBA.
Determination of repeatability.
The interassay variation measured as the coefficient of variation derived from eight runs of a panel of seven selected control samples showed that run-to-run variation for all seven samples was very low (Tables 1 and 2).
Within-assay variation of samples on the PCV2/PRRSV MBA, which was measured by placing the same sample at different locations on a plate, was also negligible. A negative sample had COV values of 0.08 and 0.04 for PRRSV and PCV2, respectively. A low-positive sample had COV values of 0.009 and 0.03 for PRRSV and PCV2, respectively. The highest COV values obtained for five other samples that were high positives for both analytes on the standard ELISAs were 0.02 and 0.07 for PRRSV and PCV2, respectively.
Kinetics of antibody responses in experimentally infected pigs.
The kinetics of antibody responses in experimentally infected pigs measured by the PCV2/PRRSV MBA were comparable to the kinetics of the responses measured by the standard ELISAs for PCV2 and PPRSV (Fig. 1). The analytical sensitivity measured by the earliest detection of specific antibodies occurred between 3 dpi and 10 dpi for PCV2 in both assays. Similarly, for PRRSV the earliest detection of a specific response was between 3 dpi and 7 dpi on both assays. Thereafter, the pattern of detection of incremental increases over time points was similar for both the PCV2/PRRSV MBA and ELISAs for PCV2 and PRRSV until 21 dpi. Between 21 dpi and 28 dpi, there was a difference between the PCV2/PRRSV MBA and ELISAs in the PRRSV antibody detection patterns. The PRRSV antibody response appeared to show an increasing trend in the ELISA, while it had reached a plateau and was decreasing in the PCV2/PRRSV MBA. The combined detection of PCV2 and PRRSV antibodies in coinfected animals was not affected by multiplexing, as the trend lines for the dually infected animals remained similar for the gold standard ELISAs and the PCV2/PRRSV MBA. Samples from mock-infected animals and samples from 1 dpi remained negative on both assays. The median blank fluorescence intensity values measured in wells in which the primary antibody was omitted were negligible (data not shown).
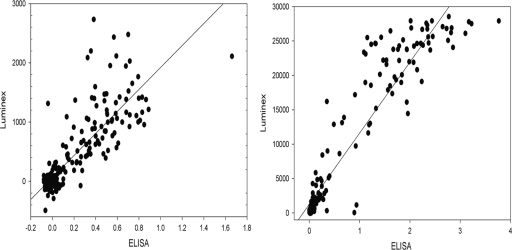
Good agreement was observed between the PCV 2 and PRRSV ELISAs and PCV2/PRRSV MBA at 7 dpi, which is considered a critical time point for determining seroconversion for PRRSV (Spearman's rank correlation = 0.72, P < 0.001). Similarly, for PCV2 the Spearman's rank correlation was 0.80 (P < 0.001) at 14 dpi. This time point was used for evaluation as seroconversion is usually slower for PCV2. When the S/P ratios of individual samples measured by the standard ELISAs were plotted against the median fluorescence units obtained from the PCV2/PRRSV MBA, the relationship was found to be linear at 7, 14, and 21 dpi, with the plots for a majority of samples clustering around the trend line (Fig. 2).
Fig. 2.
Scatter plots showing the linear relationship between the gold standard ELISAs and the PCV/PRRSV MBA. Data depicted are for samples collected from pigs that were experimentally infected with PCV2 or PRRSV, or both. PCV2 ELISA OD values (x axis) and the PCV2 MBA median fluorescence intensity values (Luminex) (y axis) are shown on the left panel. PRRSV ELISA OD values (x axis) and the PPRRSV MBA median fluorescence intensity values (Luminex) (y axis) are shown on the right panel.
We had previously determined that antibody responses in dually infected animals were not significantly different from those in singly infected animals at 21 and 28 dpi using the standard ELISA (29). The PCV2/PRRSV MBA also showed that PCV2 and PRRSV antibody responses in singly infected pigs were not different from those in the dually infected pigs at 21 and 28 dpi (Wilcoxon's rank sum test, P > 0.05), indicating that multiplexing did not interfere with detection of either analyte.
ROC analysis for determination of cutoff limits.
To avoid compromising assay sensitivity or specificity by selecting cutoff values based exclusively on negative samples, detailed ROC analysis, which takes into account both the positive and the negative reference populations in the data set, was carried out. The areas under the curve (AUCs) were 0.97 for PCV2 and 0.92 for PRRSV. The positive and negative cutoff values selected on the basis of the ROC analysis results for PCV2 were <200 MFU for negative samples, 200 to 400 for suspect samples, and >400 MFU for positive samples. The diagnostic sensitivity and specificity were both 98% for the PCV2 MBA. For the PRRSV MBA, the cutoff values were <4,000 MFU for negative samples, 4,000 to 7,000 for suspect samples, and >7,000 MFU for positive samples. The diagnostic sensitivity and specificity were 93% and 91%, respectively, for the PRRSV MBA. On the basis of optimal sensitivity and specificity values, the positive/negative cutoff values were selected to be 400 MFU for PCV2 and 7,000 MFU for PRRSV. The kappa statistic values, which represent agreement with the standard ELISAs, were 0.79 (95% confidence interval [CI], 0.74 to 0.84) and 0.67 (95% CI, 0.60 to 0.74) for the PCV2 and PRRSV MBAs, respectively. At these cutoffs, the diagnostic sensitivity and specificity for the PCV2 MBA were both 98%, while they were 93 and 91%, respectively, for the PRRSV MBA. On the basis of the results for the field samples, the positive and negative predictive values for the PCV2 MBA were both 98% and the accuracy of the test was 92%. The positive and negative predictive values for the PRRSV MBA were 89% and 73%, respectively, and the accuracy of the test was 82%.
Validation of PCV2/PRRSV MBA with field samples.
To validate the PCV2 MBA, 388 field samples were assessed by the standard PCV2 ELISA and the PCV2/PRRSV MBA. A comparison of the distribution of negative, suspect, and positive samples measured by the PCV2/PRRSV MBA and gold standard ELISA is shown in Fig. 3. While 226 of the samples were classified negative on the basis of the ELISA, 240 samples were classified negative on the basis of the PCV2/PRRSV MBA (Fig. 3). One hundred thirty-four samples were classified positive on the basis of the ELISA, while 105 of these samples were classified positive on the basis of the PCV2/PRRSV MBA. Twenty-eight samples were classified suspect using the ELISA, while 43 samples were classified suspect on the basis of the PCV2/PRRSV MBA cutoffs.
The dynamic range of the PCV2/PRRSV MBA was assessed by selecting samples that ranged from low positives (S/P ≥ 0.3) to high positives (S/P ≤ 3.1) on the standard ELISA. The two assays were comparable, as the average MFU values for field samples that were classified negative by the standard PCV2 ELISA were also low on the PCV2/PRRSV MBA (50.17 ± 110.11), while the average MFU value for the suspect samples was 292.59 ± 66.33. The highest MFU recorded with the PCV2/PRRSV MBA for the positive samples was 5,702, with a mean value of 1,120.
Similarly, 378 field samples that included low to high positives on the PPRSV ELISA were analyzed by the PCV2/PRRSV MBA. On the basis of the cutoff of the commercial ELISAs, 262 of the analyzed samples were classified negative and the remaining 116 samples ranged from low to high positive. On the basis of the PCV2/PRRSV MBA, 213 samples were classified negative, 51 suspect, and 114 positive. The dynamic ranges for both assays were similar, as the average of the MFU values for the negative samples was 3,335.56 (±5,421.40), while the median value was 1,425.00. The average MFU value for positive samples was 12,680.48 (±7,796.72), and the median value was 11,181.00.
Of the 130 samples that were analyzed by both the PCV2 and PRRSV standard ELISA, samples that were negative on the ELISAs had very low MFU values on the PCV2/PRRSV MBA as well. Samples that were positive for both analytes on the standard ELISAs were also positive on the PCV2/PRRSV MBA, with average MFU values of 1,189.83 and 2,2700.67 for PCV2 and PRRSV, respectively. Therefore, data generated from field samples reconfirmed conclusions based on the experimental samples; i.e., multiplexing did not interfere with detection of either analyte.
DISCUSSION
Unlike human medicine, the detection of antibodies to multiple pathogens from a single test is of particular value in veterinary medicine, where herd health monitoring relies on statistics-based sampling of a small proportion of the total population to minimize cost. Rarely are entire swine herds screened to monitor for selected pathogens (26). Multiplex serology may also be of special value to determine the optimal timing of multivalent vaccine administration after waning of a maternal antibody response and to monitor the duration of vaccinal immunity (16, 35). Multiplex serology plays an important role in clinical differential diagnosis and does so more for economically important swine diseases where multiple antigenic subtypes or serotypes are involved, e.g., SIV infection, leptospirosis, and A. pleuropneumoniae infection (24). To the best of our knowledge, this study is the first to describe an advanced multiplex antibody detection assay for economically important swine viruses.
The performance of our PCV2/PRRSV MBA was similar to that of the gold standard ELISAs in sensitivity of detection of both PCV2 and PRRSV in sera with experimentally generated viral infections. However, at later time points, where the magnitude of the antibody response is higher, there were differences in the patterns of detection between the standard tests and the PCV2/PRRSV MBA. These findings indicate that the dynamic ranges of the two assays differed at the higher end. However, this difference was not statistically significant, based on Spearman's correlation coefficient values of 0.72 and 0.80 for PCV2 and PRRSV, respectively. Since the diagnostic value of serological testing is primarily to differentiate negative and positive, the practical utility of the PCV2/PRRSV MBA was comparable to that of the standard tests. In situations where the strength of the antibody response is important, a titration curve can be utilized. The PCV2 ELISA rather than the animal's presumed true-positive status based on experimental infection was used as the standard for the analysis of experimental samples because the true status of the animal at between 7 dpi and 14 dpi, when seroconversion occurs, is variable until the titers are high enough for the animal to become clearly positive. As the standard ELISA cutoffs have already been validated to accommodate such samples in the gray zone (27), the PCV2/PRRSV MBA was validated by comparison to the standard ELISA and not the presumed true-positive status of the animal.
Assessment of the diagnostic performance of the PCV2/PRRSV MBA with field samples whose status was previously determined by the standard ELISAs showed excellent agreement for the PCV2 MBA with the standard test. The PRRSV MBA had a kappa statistic value of 0.67, which indicated good agreement with the standard test. The negative predictive value for the test was 73% and the accuracy was 82%. The standard PRRSV ELISA is a categorical test that classifies samples as either positive or negative and has no assigned suspect range. The relatively high cutoff value of 0.4 was probably selected to avoid unexpected positive reactions (3, 7, 17, 30). However, having a relatively high cutoff value would lead to suspect samples being classified negative by the commercial kit, thus explaining the less than desirable negative predictive value for the MBA and the excellent agreement of the PRRSV ELISA and MBA on classification of positive samples. Moreover, the N-protein antigen used in this study was derived from North American PRRSV strain VR2332. It is known that the N proteins of the American and European-like PRRSV isolates share only about 65% identity and have some dissimilar epitopes (5, 25). Therefore, future plans to incorporate the N protein derived from a type I PRRSV isolate into the PCV2/PRRSV MBA might improve the sensitivity and assay performance for field samples where antibodies to both strains may be present. Similarly, taking into account the emergence of PCV type 2b in U.S. swine populations, addition of the PCV2b capsid protein to the assay may improve the umbrella of detection (2, 31). The nsp7 protein of PRRSV has been reported to be successful in distinguishing unexpected positive samples (4). To improve the sensitivity and specificity of detection by the PCV2/PRRSV MBA, we tested the PRRSV nsp2p protein's ability to distinguish the unexpected false-positive samples. However, no clear differential recognition patterns were obvious (unpublished data).
Selecting a single arbitrary cutoff value entirely on the basis of negative samples could result in a loss of sensitivity and increased assay variation (8). Therefore, the nonparametric ROC analysis, which is based on both the positive and the negative reference samples, was used to calculate cutoffs to distinguish between positive and negative samples (9, 10). The specificity was plotted against sensitivity for the PCV2/PRRSV MBA at multiple AFU values. The working assay cutoffs were selected by comparison of the best sensitivity and specificity values of multiple AFUs. The area under the curve, which is a measure of the diagnostic accuracy of the test (12), was acceptable for both the PRRSV and PCV2 MBAs at values of 0.97 and 0.92, respectively. While the negative predictive value of the PRRSV MBA could have been improved by lowering the cutoff value of 7,000 AFU, the selected cutoff was preferred to retain specificity even at the cost of sensitivity. Although the commercial PRRSV ELISA does not have a suspect range, to avoid any possible error in classifying samples positive or negative, a suspect range was selected for the PCV2/PRRSV MBA. The status of samples that fall in this zone should be confirmed by additional testing.
In summary, we have developed and validated an advanced method for simultaneous detection of antibody responses to two important swine pathogens. This assay requires a smaller sample volume and less time than conventional immunoassays and is more economical and easy to use than two individual immunoassays. Increasing the multiplexing capability of this MBA will result in a high-throughput, flexible, and sensitive platform with practical diagnostic, epidemiological, and disease surveillance utilities.
ACKNOWLEDGMENTS
This study was funded by the Department of Veterinary Diagnostic and Production Animal Medicine, College of Veterinary Medicine, Iowa State University.
We acknowledge Susan Wong for advice on assay development. We also thank X. J. Meng, K. J. Yoon, J. J. Zimmerman, P. G. Halbur, and the staff of the Serology Section at the Veterinary Diagnostic Laboratory, Department of Veterinary Diagnostic and Production Animal Medicine, College of Veterinary Medicine, Iowa State University, for their input and help with locating and obtaining resources. Bruce Janke provided the SIV-specific serum samples used in this study. We also thank M. Hines, M. Coarsey, and D. Ingram for reviewing the manuscript.
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Balasuriya U. B., et al. 2006. Detection of antibodies to West Nile virus in equine sera using microsphere immunoassay. J. Vet. Diagn. Invest. 18: 392–395 [DOI] [PubMed] [Google Scholar]
- 2. Beach N. M., Ramamoorthy S., Opriessnig T., Wu S. Q., Meng X. J. 2010. Novel chimeric porcine circovirus (PCV) with the capsid gene of the emerging PCV2b subtype cloned in the genomic backbone of the non-pathogenic PCV1 is attenuated in vivo and induces protective and cross-protective immunity against PCV2b and PCV2a subtypes in pigs. Vaccine 29: 221–232 [DOI] [PubMed] [Google Scholar]
- 3. Brockmeier S. L., Halibur P. G., Thacker E. L. 2002. Porcine respiratory disease complex, p. 231–259 In Brogden K. A., Guthmiller J. M. (ed.), Polymicrobial diseases. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 4. Brown E., et al. 2009. Antibody response to porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural proteins and implications for diagnostic detection and differentiation of PRRSV types I and II. Clin. Vaccine Immunol. 16: 628–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dea S., Gagnon C. A., Mardassi H., Pirzadeh B., Rogan D. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 145: 659–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fenaux M., et al. 2002. Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: characterization of clinical disease, virus distribution, and pathologic lesions. J. Virol. 76: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferrin N. H., et al. 2004. Validation of a blocking enzyme-linked immunosorbent assay for detection of antibodies against porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 11: 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Go Y. Y., et al. 2008. Development of a fluorescent-microsphere immunoassay for detection of antibodies specific to equine arteritis virus and comparison with the virus neutralization test. Clin. Vaccine Immunol. 15: 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Greiner M. 1995. Two-graph receiver operating characteristic (TG-ROC): a Microsoft-Excel template for the selection of cut-off values in diagnostic tests. J. Immunol. Methods 185: 145–146 [DOI] [PubMed] [Google Scholar]
- 10. Greiner M. 1996. Two-graph receiver operating characteristic (TG-ROC): update version supports optimisation of cut-off values that minimise overall misclassification costs. J. Immunol. Methods 191: 93–94 [DOI] [PubMed] [Google Scholar]
- 11. Greiner M., Gardner I. A. 2000. Application of diagnostic tests in veterinary epidemiologic studies. Prev. Vet. Med. 45: 43–59 [DOI] [PubMed] [Google Scholar]
- 12. Greiner M., Pfeiffer D., Smith R. D. 2000. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 45: 23–41 [DOI] [PubMed] [Google Scholar]
- 13. Griffin S. M., Chen I. M., Fout G. S., Wade T. J., Egorov A. I. 2011. Development of a multiplex microsphere immunoassay for the quantitation of salivary antibody responses to selected waterborne pathogens. J. Immunol. Methods 364: 83–93 [DOI] [PubMed] [Google Scholar]
- 14. Hansen M. S., et al. 2010. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J. Comp. Pathol. 143: 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He J., et al. 2009. Rapid multiplex reverse transcription-PCR typing of influenza A and B virus, and subtyping of influenza A virus into H1, 2, 3, 5, 7, 9, N1 (human), N1 (animal), N2, and N7, including typing of novel swine origin influenza A (H1N1) virus, during the 2009 outbreak in Milwaukee, Wisconsin. J. Clin. Microbiol. 47: 2772–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horlen K. P., et al. 2008. A field evaluation of mortality rate and growth performance in pigs vaccinated against porcine circovirus type 2. J. Am. Vet. Med. Assoc. 232: 906–912 [DOI] [PubMed] [Google Scholar]
- 17. Horter D. C., et al. 2002. Characterization of the carrier state in porcine reproductive and respiratory syndrome virus infection. Vet. Microbiol. 86: 213–228 [DOI] [PubMed] [Google Scholar]
- 18. Janke B. H. 1995. Diagnosis of viral respiratory diseases in swine, p. 116–120 In Swine health and production, vol. 3 American Association of Swine Veterinarians, Perry, IA [Google Scholar]
- 19. Johnson C. R., Yu W., Murtaugh M. P. 2007. Cross-reactive antibody responses to nsp1 and nsp2 of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 88: 1184–1195 [DOI] [PubMed] [Google Scholar]
- 20. Kim J., Chung H. K., Chae C. 2003. Association of porcine circovirus 2 with porcine respiratory disease complex. Vet. J. 166: 251–256 [DOI] [PubMed] [Google Scholar]
- 21. Lawson S., et al. 2010. Development of an 8-plex Luminex assay to detect swine cytokines for vaccine development: assessment of immunity after porcine reproductive and respiratory syndrome virus (PRRSV) vaccination. Vaccine 28: 5356–5364 [DOI] [PubMed] [Google Scholar]
- 22. Leuwerke B., Kitikoon P., Evans R., Thacker E. 2008. Comparison of three serological assays to determine the cross-reactivity of antibodies from eight genetically diverse U.S. swine influenza viruses. J. Vet. Diagn. Invest. 20: 426–432 [DOI] [PubMed] [Google Scholar]
- 23. Lindau-Shepard B. A., Pass K. A. 2010. Newborn screening for cystic fibrosis by use of a multiplex immunoassay. Clin. Chem. 56: 445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacInnes J. I., et al. 2008. Prevalence of Actinobacillus pleuropneumoniae, Actinobacillus suis, Haemophilus parasuis, Pasteurella multocida, and Streptococcus suis in representative Ontario swine herds. Can. J. Vet. Res. 72: 242–248 [PMC free article] [PubMed] [Google Scholar]
- 25. Meng X. J., Paul P. S., Halbur P. G., Lum M. A. 1995. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch. Virol. 140: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mortensen S., Strandbygaard B., Botner A., Feld N., Willeberg P. 2001. Monitoring porcine reproductive and respiratory syndrome virus infection status in swine herds based on analysis of antibodies in meat juice samples. Vet. Res. 32: 441–453 [DOI] [PubMed] [Google Scholar]
- 27. Nawagitgul P., et al. 2002. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin. Diagn. Lab. Immunol. 9: 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nolan J. P., Mandy F. F. 2001. Suspension array technology: new tools for gene and protein analysis. Cell. Mol. Biol. (Noisy-le-Grand, France) 47: 1241–1256 [PubMed] [Google Scholar]
- 29. Ogawa H., et al. 2009. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J. Virol. Methods 160: 210–214 [DOI] [PubMed] [Google Scholar]
- 30. Okinaga T., et al. 2009. Evaluation of unexpected positive results from a commercial ELISA for antibodies to PRRSV. Vet. Rec. 164: 455–459 [DOI] [PubMed] [Google Scholar]
- 31. Opriessnig T., et al. 2008. Differences in virulence among porcine circovirus type 2 isolates are unrelated to cluster type 2a or 2b and prior infection provides heterologous protection. J. Gen. Virol. 89: 2482–2491 [DOI] [PubMed] [Google Scholar]
- 32. Ramamoorthy S., Huang F. F., Huang Y. W., Meng X. J. 2009. Interferon-mediated enhancement of in vitro replication of porcine circovirus type 2 is influenced by an interferon-stimulated response element in the PCV2 genome. Virus Res. 145: 236–243 [DOI] [PubMed] [Google Scholar]
- 33. Ramamoorthy S., Opriessnig T., Pal N., Huang F. F., Meng X. J. 2011. Effect of an interferon-stimulated response element (ISRE) mutant of porcine circovirus type 2 (PCV2) on PCV2-induced pathological lesions in a porcine reproductive and respiratory syndrome virus (PRRSV) co-infection model. Vet. Microbiol. 147: 49–58 [DOI] [PubMed] [Google Scholar]
- 34. Shoma S., et al. 2011. Development of a multiplexed bead-based immunoassay for the simultaneous detection of antibodies to 17 pneumococcal proteins. Eur. J. Clin. Microbiol. Infect. Dis. 30: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thacker E., Janke B. 2008. Swine influenza virus: zoonotic potential and vaccination strategies for the control of avian and swine influenzas. J. Infect. Dis. 197(Suppl. 1): S19–S24 [DOI] [PubMed] [Google Scholar]
- 36. Thacker E. L. 2001. Immunology of the porcine respiratory disease complex. Vet. Clin. North Am. Food Anim. Pract. 17: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vincent A. L., et al. 2010. Experimental inoculation of pigs with pandemic H1N1 2009 virus and HI cross-reactivity with contemporary swine influenza virus antisera. Influenza Other Respir. Viruses 4: 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yue F., Cui S., Zhang C., Yoon K. J. 2009. A multiplex PCR for rapid and simultaneous detection of porcine circovirus type 2, porcine parvovirus, porcine pseudorabies virus, and porcine reproductive and respiratory syndrome virus in clinical specimens. Virus Genes 38: 392–397 [DOI] [PubMed] [Google Scholar]