Abstract
Helicobacter pylori infection is common in Alaska. The development of severe H. pylori disease is partially determined by the virulence of the infecting strain. Here we present vacA and cagA genotype data for H. pylori strains isolated from Alaskans and their correlation with clinical disease. We enrolled patients scheduled for esophagogastroduodenoscopy and positive for H. pylori infection. Gastric biopsy specimens from the stomach antrum and fundus were cultured. We performed PCR analysis of the H. pylori vacA gene and for the presence of the cagA gene and cagA empty site. We genotyped 515 H. pylori samples from 220 Native and 66 non-Native Alaskans. We detected the cagA gene in 242/286 (85%) persons; of 222 strains that could be subtyped, 95% (212) were non-Asian cagA and 3% (6) were East Asian cagA. After removing mixed infections (n = 17), 83% of H. pylori strains had either the vacA s1m1 (120/269) or s2m2 (103/269) genotype. Sixty-six percent (68/103) of H. pylori strains with the vacA s2m2 genotype also contained the cagA gene. Infection with an H. pylori strain having the cagA gene or vacA s1m1 genotype (compared with s1m2 and s2m2) was associated with a decreased risk of esophagitis (P = 0.003 and 0.0003, respectively). Infection with an H. pylori strain having the vacA s1m1 genotype (compared with s1m2 and s2m2) was associated with an increased risk of peptic ulcer disease (PUD) (P = 0.003). The majority of H. pylori strains in this study carried the non-Asian cagA gene and either the vacA s1m1 or s2m2 genotype. A majority of H. pylori strains with the vacA s2m2 genotype also contained the cagA gene. There was an association of H. pylori genotype with esophagitis and PUD.
INTRODUCTION
Helicobacter pylori is one of the most common infections of humans, with over 80% of persons infected in some developing countries (5). Persons infected with H. pylori have mild to severe gastric mucosal inflammation, but in some people infection leads to peptic ulcer disease (PUD) (28). Additionally, H. pylori-infected persons have at least a 2-fold increased risk of developing gastric cancer compared with uninfected persons, and H. pylori is characterized by the World Health Organization as a class 1 carcinogen and a risk factor for non-cardia gastric adenocarcinoma (18, 45). It is estimated that persons infected with this organism have a 10 to 20% lifetime risk of developing PUD and a 1 to 2% lifetime risk of developing gastric cancer (20, 21). The risk of developing these diseases depends upon the inflammatory response to chronic colonization, which is thought to be determined by a combination of factors, including the virulence of the infecting strain, the host's response to infection, and environmental cofactors.
Two putative bacterial markers of virulence are the cytotoxin-associated gene pathogenicity island (cag PAI) and the vacuolating cytotoxin gene (vacA). The cagA gene, which codes for a 125- to 145-kDa protein, CagA, is a marker for the presence of the cag PAI; however, not all strains with an expressed CagA protein express the entire set of cag PAI proteins (6, 7, 12). In western populations, persons infected by H. pylori strains containing the cag PAI usually have an elevated inflammatory response and are at a higher risk of developing PUD or gastric cancer than persons infected by H. pylori strains without the cag PAI (34, 36). Because over 90% of H. pylori strains isolated from East Asia contain the cag PAI, the relationship between the cag PAI and clinical outcome is difficult to establish in Asian populations (39).
Active VacA is a toxin that induces massive vacuolization in epithelial cells in vitro. Although all strains of H. pylori have a vacA gene, there is variation in the amount of vacuolating activity due to sequence heterogeneity within the vacA gene at the 5′ end (signal [s] region) and the middle (m) region. Two allelic s region types have been identified, s1 and s2. The s1 type can be further subtyped as s1a, s1b, and s1c. Two allelic m region types have been identified, m1 and m2. The m1 type can be further subtyped as m1a and m1b. In HeLa cells, the level of vacuolating activity is high in H. pylori strains with the s1m1 genotype, intermediate in those with the s1m2 genotype, and low or absent in those with the s2m2 genotype (3). Many groups have found a correlation between toxin activity and the pathogenicity of H. pylori; the vacA s1m1 genotype is often associated with PUD and gastric cancer in western populations and some multiregional studies (3, 33, 40). The vacA s1m1 genotype is often linked with the presence of the cag PAI, and the vacA s2m2 genotype is usually linked with the absence of the cag PAI (3).
H. pylori infection is common in Alaska. In a statewide survey of over 2,000 samples of blood collected in the 1980s, 75% of Alaska Native persons were positive for antibodies to H. pylori, indicating a current or past infection (35). The high prevalence of H. pylori in Alaska, along with high levels of antimicrobial resistance (8–10), makes it difficult to test and treat all infected persons. Therefore, it is important to identify those persons at a higher risk for severe disease to allow optimal clinical treatment and intensive follow-up. In addition, the determination of the genotype of H. pylori isolates from this population may allow us to further understand the relationship between putative virulence genes and clinical disease. We present here the first large investigation of the vacA and cagA genotypes from H. pylori strains isolated in different ethnic groups in Alaska and their correlation with clinical disease.
MATERIALS AND METHODS
Participants.
Participants in this study were a subset of those recruited as part of a previously described study of H. pylori reinfection conducted from September 1998 through January 2005 (29). Briefly, patients ≥18 years of age undergoing esophagogastroduodenoscopy (EGD) were recruited at three urban medical facilities in Anchorage, AK, and three rural medical facilities in western Alaska. Patients were eligible for enrollment into the parent study if they had a positive [13C]urea breath test (UBT; Meretek Diagnostics Inc., Lafayette, CO) at the time of their EGD. Persons were eligible for this study if their H. pylori infection was also confirmed by culture. Persons were excluded from study consideration if they were pregnant, had a prior gastric resection, had gastric cancer, or were undergoing active chemotherapy. There were two groups of patients: (i) Alaska Native persons (the indigenous peoples of Alaska) living in Anchorage or one of three regions in western Alaska and (ii) non-Native persons (nonindigenous people residing in Alaska) living in Anchorage. At enrollment, a research nurse interviewed participants by using a standardized form to obtain demographic data and information about symptoms of abdominal discomfort. A medical chart review was conducted to determine if there was a history of chronic stomach problems, specifically PUD, gastritis, or gastric surgery. Clinical diagnoses of PUD, gastritis, duodenitis, and esophagitis were determined by endoscopic evaluation using a scoring system described previously (29). All endoscopists went through extensive training to ensure that patients were evaluated in a standardized manner.
The study was approved by the Centers for Disease Control and Prevention, the Alaska Area, and the Western Institutional Review Boards as well as the Alaska Native Tribal Health Consortium and four tribal health boards: Southcentral Foundation, Yukon-Kuskokwim Health Corporation, Bristol Bay Native Health Corporation, and Norton Sound Health Corporation. Study participants provided written informed consent.
H. pylori isolates and DNA extraction.
During the EGD, gastric biopsy specimens were collected from the antrum and/or fundus of the stomach. The participant's endoscopist determined how many biopsy specimens were collected and from where they were collected based upon the clinical needs of the patient. Biopsy specimens were cultured as previously described (30), and all H. pylori organisms were stored at −80°C. At the time of DNA extraction, stored H. pylori organisms were cultured on Trypticase soy agar plates containing 5% sheep blood (Remel, Lenexa, KS) and incubated for 3 to 5 days at 37°C in 12% CO2 and high humidity. H. pylori cells from each plate were collected together by sweeping a 1-μl inoculating loop across the medium. The H. pylori cells were placed into 100 μl of phosphate-buffered saline and vortexed to create a uniform turbidity. DNA was extracted with the MagNA Pure Compact using MagNA Pure Compact nucleic acid isolation kit I and the DNA Cultured Cells v3.1 protocol (Roche Applied Science, Indianapolis, IN). This protocol uses proteinase K and a chaotropic salt-containing lysis buffer to lyse the cells and magnetic glass particles to collect the nucleic acids.
PCR primers and probes.
PCR analysis was performed on H. pylori DNA samples to genotype the vacA s and m regions and to detect the presence of the cagA gene and the cagA empty site using previously described primers (Table 1). Real-time PCR probes were designed from sequences obtained from the National Center for Biotechnology Information database by using Primer3 software (Table 1). Probes were tested for potential cross-reactivity with nonhomologous viral and bacterial sequences by using the Basic Local Alignment Search Tool software and were tested for specificity by using DNA collected from various Helicobacter, Campylobacter, and Wolinella species.
Table 1.
PCR primer and probe sequences for amplification of the cagA and vacA genes
| Region | Primer or probe | Oligonucleotide sequence | Reference |
|---|---|---|---|
| cagA, East Asian | CAG1F | 5′-TGG AAC CCT AGT CGG TAA TGG G | 48 |
| CAG1R | 5′-TGA TGC AAT TTT GTT AAT CCG GTC | 48 | |
| CAG1P | 5′-TCT AAA ACA GAA GCC ACA ACG CTC ACC | ||
| cagA, non-Asian | CAG2F | 5′-AAT GCA AAA ATT GAC CRA CTC AAT C | 48 |
| CAG2R | 5′-AAA CCT GCT TTA GCT TCT GAY ACY GC | 48 | |
| CAG2P | 5′-CTT CCC TTT GAA AAG GCA TGA TAA AGT TG | ||
| cagA, 5′ end | CAGF | 5′-AAT GGT GGT CCT GGA GCT AG | 32 |
| CAGR | 5′-GGA AAT CTT TAA TCT CAG TTC GGA A | 32 | |
| cag PAI empty site | Luni1 | 5′-CA TTT TGG CTA AAT AAA CGC TG | 32 |
| r5280 M | 5′-TTG CAC GCA TTT TCC CTT AAT C | 32 | |
| vacA s1a | SS1-F | 5′-GTC AGC ATC ACA CCG CAA C | 3 |
| VAI-R | 5′-CTG CTT GAA TGC GCC AAA C | 3 | |
| S1aP | 5′-AAA CAA GCC GAA GAA GCC AAT AAA ACC CC | ||
| vacA s1b | SS3-F | 5′-AGC GCC ATA CCG CAA GAG | 3 |
| VAI-R | 5′-CTG CTT GAA TGC GCC AAA C | 3 | |
| vacA s1c | S1c-F | 5′-CTY GCT TTA GTR GGG YTA | 46 |
| VAI-R | 5′-CTG CTT GAA TGC GCC AAA C | 3 | |
| vacA s2 | SS2-F | 5′-GCT AAC ACG CCA AAT GAT CC | 3 |
| VAI-R | 5′-CTG CTT GAA TGC GCC AAA C | 3 | |
| S2P | 5′-ACA ACY GTG ATY ATT CCA GCC ATT GTT GG | ||
| vacA m1a | VA3-F | 5′-GGT CAA AAT GCG GTC ATG G | 3 |
| VA3-R | 5′-CTA ATG CCA TTG GTA CCT GTA GAA AC | 3 | |
| M1aP | 5′-YGT AGG CAA TGC AGC AGC TAT GAT GTT TA | ||
| vacA m1b | VAM-F3 | 5′-CCC CAA TGC AGT CAT GGA T | 32 |
| VAM-R3 | 5′-GCT GTT AGT GCC TAA AGA AGC AT | 32 | |
| M1bP | 5′-CAC TAT CAA TTA TTT GGT TCG AGG CGG GAA | ||
| vacA m2 | VA4-F | 5′-GGA GCC CCA GGA AAC ATT G | 3 |
| VA4-R | 5′-TGT CAT AAC TAG CGC CTT GCA C | 3 | |
| M2P | 5′-CTT TTT GTC CAA GAT GGG CGT GTA GC |
cagA genotyping.
All samples were initially tested with the Smartcycler real-time detection system (Cephid, Sunnyvale, CA), using Omnimix beads and fluorescent probes (Table 1) to amplify the 3′ end of the cagA gene and identify either the East Asian or non-Asian cagA gene (48). Samples negative by these two reactions were tested with a Stratagene MX3000 instrument using the Stratagene Brilliant reaction mixture (Stratagene, La Jolla, CA) to detect the 5′ end of the cagA gene (32). Products from this reaction were viewed on a 1% agarose gel. Samples positive for the cagA 5′ reaction were tested once more to delineate between the East Asian and non-Asian cagA genes; however, this time, a conventional detection system (1% agarose gel), rather than a real-time system, was used to view the products.
To detect the cag PAI empty site (indicating the absence of the cag PAI) (32), samples were tested with an Applied Biosystems 9700 thermal cycler using Applied Biosystems 10× PCR buffer (Applied Biosystems Inc., Foster City, CA). Products were viewed on a 1% agarose gel.
vacA genotyping.
All samples were tested with the Stratagene MX3000 real-time detection system using the Stratagene Brilliant quantitative PCR (QPCR) mixture (Stratagene, La Jolla, CA) and previously described primers (Table 1) (3, 32, 46). Products from the vacA s1b and s1c subtypes were viewed on a 3% agarose gel. All other vacA allelic types and subtypes were detected by using fluorescent probes (Table 1). To confirm the results generated by PCR, the vacA s and m regions from a subset of samples were sequenced on an Applied Biosystems 3130 genetic analyzer (Applied Biosystems Inc., Foster City, CA) according to previously described methods (3, 4).
Statistical analysis.
For all statistical comparisons, persons infected by H. pylori strains with mixed vacA allelic types (s1/s2 or m1/m2) or incomplete vacA genotypes (the m region or s region could not be typed) were removed. Persons with mixed allelic subtypes (i.e., s1a/s1b or m1a/m1b) were considered to be s1 and m1, respectively, and were not excluded from the analysis. Mixed vacA infections occurred because (i) H. pylori strains cultured from the antrum and fundus of one participant had different genotypes or (ii) H. pylori strains cultured from a single site (antrum or fundus) contained more than one genotype. Unless stated otherwise, persons infected with both cagA-positive and cagA-negative colonies of H. pylori were considered to be infected with a cagA-positive strain. The likelihood ratio χ2 test was used to test for differences in the two groups (Alaska Native and non-Native persons), and P values were exact when the sample size necessitated.
For the correlation of H. pylori genotypes with clinical data, we restricted the evaluation to three combinations of vacA s and vacA m regions with adequate sample sizes for evaluation: s1m1, s1m2, and s2m2. We compared clinical variables between these three genotypes with the likelihood ratio χ2 test and analysis of variance (ANOVA) for categorical and continuous variables, respectively. All P values are two sided, and a P value of <0.05 was considered statistically significant.
RESULTS
Study participants.
Genotyping was performed on 515 H. pylori samples collected from 220 Alaska Native and 66 non-Native persons (Table 2). Sixty-eight participants (24%) had one sample available (from either the antrum or fundus), 211 participants (74%) had two samples available (from both the antrum and fundus), and 7 participants (2%) had three or more samples available (more than one sample from the antrum and/or fundus). Endoscopic reports were available for 260 (91%) of the 286 participants. The most common clinical diagnoses were mild to moderate gastritis (198/260 [76%]), followed by esophagitis (63/260 [24%]). Fifty-eight percent (152/260) of participants had either a healthy stomach (33/260 [13%]) or gastritis only (119/260 [46%]). All Alaska Native persons and 68% (45/66) of non-Native persons were born in the United States.
Table 2.
Study participant demographics and clinical diagnoses, Alaska, 1998 to 2005
| Characteristic | Value |
|---|---|
| Demographics (n = 286) | |
| Mean age (yr) (min, max) | 49 (18, 88) |
| No. (%) of patients | |
| Female | 173 (60) |
| Alaska Native | 220 (77) |
| Place of birth | |
| United States | 265 (93) |
| Asia | 9 (3) |
| Non-U.S. Americas | 7 (2) |
| Europe | 4 (1) |
| Middle East | 1 (0.3) |
| No. (%) of patients taking medications (n = 286) | |
| NSAIDa | 34 (12) |
| PPIb | 81 (28) |
| H2 blockerc | 113 (40) |
| Heavy alcohold | 14 (5) |
| Use of ≥1 of the above medications | 196 (69) |
| No. (%) of patients with medical history (n = 286) of: | |
| Gastritis (5 yr) | 110 (38) |
| Peptic ulcer disease | 38 (13) |
| No. (%) of patients with endoscopic evaluation resulte (n = 260) of: | |
| Duodenitis | 46 (18) |
| Esophagitis | 63 (24) |
| Ulcer | 23 (9) |
| Gastric ulcer | 18 (7) |
| Gastritis | |
| Mild | 103 (40) |
| Moderate | 95 (37) |
| Severe | 20 (8) |
NSAID, nonsteroidal anti-inflammatory drugs (daily use).
PPI, protein pump inhibitor (use within 30 days of enrollment).
Use within 30 days of enrollment.
At least 3 drinks daily.
Thirty-three (13%) persons had healthy stomachs; 100 (38%) persons had more than one diagnosis.
cagA genotyping.
The cagA gene was detected in H. pylori isolates from 242/286 (85%) participants (Table 3). The cagA empty site (indicating the absence of the cag PAI) was detected in H. pylori strains from 99/286 (35%) participants. Among the 99 participants with the cagA empty site detected, 55 (56%) also had the cagA gene, and 44 (44%) were positive for the cagA empty site only. Alaska Native persons were more likely than non-Native persons to be infected with H. pylori containing the cagA gene (198/220 [90%] versus 44/66 [67%]; P < 0.0001). The cagA genes from 222/242 (92%) participants were further characterized. A total of 212/222 (95%) participants had a non-Asian cagA-positive strain, 6/222 (3%) participants had an East Asian cagA-positive strain, and 4/222 (2%) participants had both non-Asian and East Asian cagA-positive strains.
Table 3.
cagA and vacA genotypes of Helicobacter pylori strains isolated from Alaska Native and non-Native persons in Alaska, 1998 to 2005
| Genotype | No. (%) of patients with genotype |
||
|---|---|---|---|
| Alaska Native (n = 220) | Non-Native (n = 66) | Total (n = 286) | |
| cagA gene | |||
| Absenta | 22 (10) | 22 (33) | 44 (15) |
| Presentb | 198 (90) | 44 (67) | 242 (85) |
| vacA s region type | |||
| s1a | 108 (49) | 22 (33) | 130 (45) |
| s1b | 14 (6) | 17 (26) | 31 (11) |
| s1c | 0 (0) | 6 (9) | 6 (2) |
| s2 | 86 (39) | 19 (29) | 105 (37) |
| Mixedc | 12 (5) | 2 (3) | 14 (5) |
| vacA m region type | |||
| m1a | 55 (25) | 19 (29) | 73 (26) |
| m1b | 19 (9) | 5 (8) | 24 (8) |
| m2 | 117 (53) | 40 (61) | 157 (55) |
| Mixedc | 29 (13) | 3 (5) | 32 (11) |
Only the cagA empty site is present.
Includes isolates from persons where both the cagA gene and the cagA empty site are present.
More than one allelic type/subtype is present in the same participant, suggesting infection with more than one H. pylori strain.
vacA genotyping.
The vacA s1a subtype was identified in H. pylori strains from 130/286 (45%) participants, the vacA s1b subtype was identified from 31/286 (11%) participants, the vacA s1c subtype was identified from 6/286 (2%) participants, and the vacA s2 type was identified from 105/286 (37%) participants (Table 3). The remaining 14 participants had mixed H. pylori vacA s region types or subtypes (s1a/s2, n = 8; s1b/s2, n = 2; s1c/s2, n = 1; s1a/s1b, n = 1; s1a/s1c, n = 1; s1a/s1b/s2, n = 1).
DNA sequencing of the vacA s region was completed on H. pylori samples from 33% (94/286) of study participants. The samples represented a variety of vacA s region types or subtypes, as determined by PCR (s1a, n = 35; s1b, n = 12; s1c, n = 2; s2, n = 45). All 94 sequences corroborated the PCR results.
The vacA m1a subtype was identified in H. pylori isolates from 73/286 (26%) participants, the vacA m1b subtype was identified from 24/286 (8%) participants, and the vacA m2 type was identified from 157/286 (55%) participants (Table 3). The remaining 30 participants had mixed H. pylori vacA m region types or subtypes (m1a/m1b, n = 24; m1a/m2, n = 4; m1b/m2, n = 1; m1a/m1b/m2, n = 1).
DNA sequencing of the vacA m region was completed on H. pylori samples from 33% (94/286) of study participants. The samples represented a variety of vacA m region types or subtypes, as determined by PCR (m1a, n = 27; m1b, n = 6; m2, n = 61). All 94 sequences corroborated the PCR results.
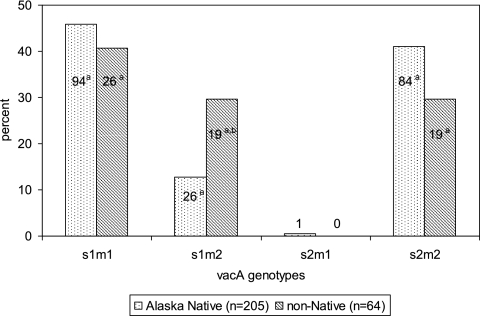
Two hundred sixty-nine participants (94%) had complete vacA genotyping results with no mixed infections. The vacA s1m1 genotype was identified in 120 (45%) participants, the vacA s1m2 genotype was identified in 48 (18%) participants, the vacA s2m1 was identified in 1 (0.4%) participant, and the vacA s2m2 genotype was identified in 103 (38%) participants (Fig. 1). Compared with the other allelic combinations, Alaska Native persons were less likely than non-Native persons to be infected with H. pylori strains having the vacA s1m2 genotype (26/205 [13%] versus 19/64 [30%]; P = 0.003).
Fig. 1.
Proportion of Helicobacter pylori vacA genotypes isolated from Alaska Native and non-Native persons in Alaska from 1998 to 2005. a, numbers within the bars represent the number of persons infected with H. pylori having that vacA genotype; b, P = 0.003 for Alaska Native people versus non-Native people.
vacA genotyping and cagA status.
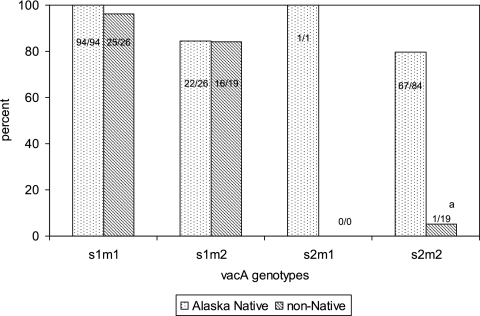
A total of 84 to 100% of H. pylori strains having the vacA s1m1 (119/120), s1m2 (38/45), and s2m1 (1/1) genotypes contained the cagA gene (Fig. 2). Sixty-six percent (68/103) of H. pylori strains with the vacA s2m2 genotype contained the cagA gene. Of H. pylori strains having the vacA s2m2 genotype, those that also contained the cagA gene were more likely to be isolated from Alaska Native persons than non-Native persons (67/84 [80%] versus 1/19 [5%]; P < 0.0001). When persons infected with both cagA-positive and cagA-negative colonies of H. pylori (n = 23 Alaska Native persons and 0 non-Native persons) were removed from the analysis, Alaska Native persons remained more likely than non-Native persons to be infected with H. pylori strains having the vacA s2m2 genotype and the cagA gene (44/61 [72%] versus 1/19 [5%]; P < 0.0001).
Fig. 2.
Percentage of Alaska Native and non-Native persons infected with a cagA-positive strain of Helicobacter pylori stratified by the vacA genotype of their infecting organism in Alaska from 1998 to 2005. Persons with both cagA-positive and cagA-negative H. pylori colonies were considered to be infected with a cagA-positive strain. a, p < 0.0001 for Alaska Native people versus non-Native people.
H. pylori genotypes and place of birth.
vacA and cagA genotyping results were stratified by the place of birth of the study participants (Table 4). All participants (6/6) infected with an East Asian cagA-positive strain of H. pylori were born in Asia, and 99% (209/212) of participants infected with a non-Asian cagA-positive strain were born in a non-Asian country. Participants born in Asia were more likely to be infected with H. pylori having the vacA m1b subtype than those born in a non-Asian country (5/8 [63%] versus 19/246 [8%]; P < 0.001). Additionally, all participants infected with a strain of H. pylori having the vacA s1c subtype (6/6) were born in Asia. The six strains of H. pylori with an East Asian cagA gene were more likely to have the vacA s1c (5/6 [83%]) and m1b (5/6 [83%]) subtypes than H. pylori strains with a non-Asian cagA gene (1/200 [0.5%] and 19/181 [10%], respectively; P < 0.001 for both).
Table 4.
cagA and vacA genotypes of Helicobacter pylori isolated from Alaska Native and non-Native persons stratified by the participants' place of birth, Alaska, 1998 to 2005
| Place of birth | No. (%) of patients with isolates of genotype |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
cagAa |
vacA s regionb |
vacA m regionb |
|||||||
| East Asian | Non-Asian | s1a | s1b | s1c | s2 | m1a | m1b | m2 | |
| United States | 0 (0) | 201 (95) | 124 (95) | 27 (87) | 0 (0) | 101 (96) | 68 (93) | 19 (79) | 147 (94) |
| Asia | 6 (100) | 3 (1) | 2 (2) | 1 (3) | 6 (100)c | 0 (0) | 1 (1) | 5 (21)c | 2 (1) |
| Non-U.S. Americas | 0 (0) | 5 (2) | 2 (2) | 3 (10) | 0 (0) | 2 (2) | 4 (5) | 0 (0) | 3 (2) |
| Europe | 0 (0) | 2 (1) | 2 (2) | 0 (0) | 0 (0) | 2 (2) | 0 (0) | 0 (0) | 4 (3) |
| Middle East | 0 (0) | 1 (0.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0.6) |
Persons with both East Asian and non-Asian cagA colony types were removed.
Persons with mixed vacA infections were removed.
All isolates also had an East Asian cagA gene.
H. pylori genotypes and clinical presentation.
Infection with H. pylori strains having the cagA gene or the vacA s1m1 genotype was associated with a decreased risk of esophagitis (Table 5). When restricting the analysis to participants with a cagA-positive strain of H. pylori, the vacA s1m1 genotype remained significantly associated with a decreased risk of esophagitis (s1m1, 15/107 [14%]; s1m2, 7/31 [23%]; s2m2, 22/67 [33%]; P = 0.01). Likewise, when controlling for the two vacA genotypes with both cagA-positive and cagA-negative strains (s1m2 and s2m2), the absence of the cagA gene remained associated with esophagitis; however, it was no longer statistically significant (cagA positive, 27/98 [28%]; cagA negative, 17/37 [46%]; P = 0.07). Infection with H. pylori having the vacA s1m1 genotype was also associated with an increased risk of having an ulcer at study enrollment or a history of PUD. This remained true after the analysis was restricted to participants with a cagA-positive strain of H. pylori (s1m1, 29/107 [27%]; s1m2, 4/31 [13%]; s2m2, 7/67 [10%]; P = 0.01). When the analysis was restricted to participants without possible confounding factors (use of nonsteroidal anti-inflammatory drugs, protein pump inhibitors, H2 blockers, and/or heavy alcohol), all trends remained the same (data not shown). Participants infected with a strain of H. pylori having the vacA s2m2 genotype did not differ in their clinical presentations based upon the presence or absence of the cagA gene. No other associations were found between H. pylori cagA and vacA genotypes and clinical diagnoses in this study population, and all associations were similar in Alaska Native and non-Native persons.
Table 5.
Clinical presentation of Alaska Native and non-Native persons infected with Helicobacter pylori strains with and without the cagA gene and with vacA genotypes s1m1, s1m2, and s2m2 in Alaska from 1998 to 2005
| Clinical presentation |
cagA statusa |
vacA genotypeb |
|||||
|---|---|---|---|---|---|---|---|
| No. (%) of patients |
P value | No. (%) of patients with genotype |
P valuec | ||||
| Positive (n = 222) | Negative (n = 38) | s1m1 (n = 107) | s1m2 (n = 37) | s2m2 (n = 98) | |||
| Endoscopic evaluation | |||||||
| Duodenitis | 40 (18) | 6 (16) | 0.74 | 22 (21) | 9 (24) | 15 (15) | 0.42 |
| Esophagitis | 46 (21) | 17 (45) | 0.003 | 15 (14) | 11 (30) | 35 (36) | 0.001d |
| Moderate or severe gastritis | 96 (43) | 19 (50) | 0.44 | 48 (45) | 19 (51) | 44 (45) | 0.77 |
| All ulcers | 20 (9) | 3 (8) | 0.82 | 12 (11) | 3 (8) | 7 (7) | 0.58 |
| Gastric ulcer | 16 (7) | 2 (5) | 0.65 | 12 (11) | 1 (3) | 4 (4) | 0.07 |
| Medical chart review | |||||||
| History of gastritis | 93 (42) | 13 (34) | 0.37 | 46 (43) | 11 (30) | 43 (44) | 0.28 |
| History of PUD | 32 (14) | 4 (11) | 0.51 | 21 (20) | 4 (11) | 9 (9) | 0.08 |
| Ulcer at enrollment or history of PUD | 46 (21) | 6 (16) | 0.47 | 29 (27) | 4 (11) | 12 (12) | 0.01e |
Persons with both cagA-positive and cagA-negative H. pylori colonies were considered to be infected with a cagA-positive strain.
vacA genotype s2m1 was left out due to the small sample size (n = 1).
The P value examines differences in clinical outcomes among the 3 vacA genotypes. Boldface type indicates significance.
P value for s1m1 compared to the other 2 genotypes, 0.0003.
P value for s1m1 compared to the other 2 genotypes, 0.003.
DISCUSSION
This paper presents the largest characterization to date of H. pylori isolates collected from persons living in Alaska. H. pylori infection is common in Alaska (35, 38), and a high proportion of these strains demonstrate antimicrobial resistance (8–10), making it difficult to test and treat all infected persons. It is therefore important to identify persons at a higher risk for disease to allow optimal clinical treatment and intensive follow-up. Previous studies suggested that the vacA genotype and the prevalence of the cagA gene vary in H. pylori isolates collected from different parts of the world and that these genotypic variations affect the clinical presentation in persons infected with the organism (17, 44). It is important, therefore, to collect and report these data from different regions of the world so that we can better understand the relationship between putative virulence genes and clinical disease. The majority of H. pylori strains in this study contained the cagA gene, and persons infected with cagA-positive H. pylori strains were less likely to have an esophagitis diagnosis than persons infected with cagA-negative H. pylori strains. Persons infected with an H. pylori strain having the vacA s1m1 genotype had a decreased risk of esophagitis and an increased risk of PUD. We also identified a large number of organisms with the vacA s2m2 genotype that also contained the cagA gene; however, the presence or absence of the cagA gene in these organisms did not affect the clinical presentation of the patient.
We detected the cagA gene in 85% of H. pylori isolates collected from our study participants. Worldwide, the presence of the cagA gene varies, from a low of 50% in some Middle Eastern countries (2) to a high of 99% in many East Asian countries (11, 23). The percentage of cagA-positive H. pylori strains found in our study is most similar to data reported from Europe and regions of North America (74% to 88%) (31, 42, 46). Eighty-three percent of participants in our study were infected with organisms having either the vacA s1m1 (45%) or s2m2 (38%) genotype. The H. pylori vacA s1m1 genotype is common worldwide, with reports of 42% to 84% of H. pylori strains from around the globe having this genotype (42). The vacA s2m2 genotype is less common, where the reported worldwide prevalences vary from 0% to 57% (19, 42). The vacA s2m2 percentage found in our study is among the highest reported, 44% in Alaska Native participants and 29% in non-Native participants. Such a high prevalence has been found in only two other small studies of organisms collected from Australia (8/24 [35%]) and North Africa (16/28 [57%]) (42).
Almost all of the H. pylori strains in our study with the vacA s1m1, s1m2, and s2m1 genotypes carried the cagA gene. Additionally, 66% of H. pylori strains with the vacA s2m2 genotype also contained the cagA gene. In contrast to our data, most studies have found an association between the low-toxin-producing vacA s2m2 genotype and less virulent cagA-negative H. pylori strains (13). With the exception of a single report of a small number of isolates from Portugal and Spain, where 57% (4/7) of the organisms with the vacA s2m2 genotype also contained the cagA gene (42), we know of no other study with such a high percentage of vacA s2m2 H. pylori strains that are also cagA positive. Almost all (66/67) of the vacA s2m2- and cagA-positive organisms in our study were isolated from Alaska Native persons. When the analysis was restricted to only Alaska Native persons, 80% of the H. pylori strains with the vacA s2m2 genotype also contained the cagA gene. We did not detect a difference in clinical presentation according to the presence or absence of the cagA gene in persons infected with H. pylori strains having the vacA s2m2 genotype, so it is unclear from our study if these strains are of clinical importance. It is possible that severe disease requires that the more virulent phenotype of both of these genes be present.
In addition to the differences noted above, we did find other significant genotypic differences between our two study groups (Alaska Native and non-Native persons). Specifically, Alaska Native persons were more likely than non-Native persons to be infected with H. pylori strains containing the cagA gene. We know of only one study investigating the prevalence of H. pylori infection in non-Native Alaskans. Those authors found an H. pylori seroprevalence of 24% in non-Native school teachers who were living in rural Alaska and were predominantly Caucasian persons born in the contiguous 48 U.S. states; testing for pathogenic genes was not performed in that study (27). In Alaska Native persons, the H. pylori seroprevalence is more than three times higher than what was found in that study (35). Documented high rates of gastric cancer in Alaska Native persons (24) may be due in part to the high prevalence of cagA-positive H. pylori strains in this population.
Many investigators have reported differences in the cagA genes in H. pylori isolates collected from persons living in East Asian versus non-Asian countries (31, 41, 48). It may be important to distinguish between these two types of cagA genes, as the CagA protein coded for by most East Asian strains appears to be more biologically active than most CagA proteins coded for by non-Asian cagA genes (16, 26), and this has been associated with an increased risk of PUD and gastric cancer (22). In our study population, 95% of persons infected with a cagA-positive strain of H. pylori that could be subtyped had a non-Asian cagA-positive strain. Only six persons had an East Asian cagA-positive strain. The six strains of H. pylori with East Asian cagA were also more likely to have the vacA s1c and m1b subtypes than H. pylori strains with non-Asian cagA. These two vacA subtypes have been linked to H. pylori isolates collected from persons living in East Asian countries and are often found in H. pylori strains having a non-Asian cagA gene (43, 46, 47). All persons infected with an East Asian cagA strain in this study were born in an East Asian country, and all but three of the persons infected with H. pylori strains having a non-Asian cagA gene were born in a non-Asian country.
Three previously published papers have reported data on the genotyping of H. pylori strains isolated from persons living in Alaska; however, the number of isolates included in those studies was small, ranging from 2 to 20 isolates per study (1, 15, 47). Two of those studies used multilocus sequence typing to group H. pylori isolates (1, 15), while the third study used a PCR methodology similar to that used in this study (47). Just as we have shown in our study, all three of those papers concluded that the majority of H. pylori strains that they tested had genes that grouped them with H. pylori strains of non-Asian origin.
We found that persons infected with a cagA-positive H. pylori strain were less likely than persons infected with a cagA-negative H. pylori strain to have a diagnosis of esophagitis upon endoscopic evaluation. Although it is not universal, many other researchers have seen this same relationship, suggesting a protective role for cagA-positive strains of H. pylori (25, 37). Persons infected with H. pylori isolates having the vacA s1m1 genotype were also less likely to have an esophagitis diagnosis than those infected with an isolate having either the vacA s1m2 or s2m2 genotype. We continued to see this association even when we restricted the analysis to only cagA-positive H. pylori strains, indicating that the vacA s1m1 genotype is independently negatively associated with esophagitis in our study population. It is biologically plausible that an H. pylori strain with the vacA s1m1 genotype would be protective against esophagitis because of its high toxin activity, and we hypothesize that H. pylori strains having the more virulent vacA s1m1 genotype could increase gastric atrophy and decrease acid production and the development of esophagitis.
Persons infected with H. pylori strains having the vacA s1m1 genotype were also more likely to have an ulcer at enrollment or a history of PUD than those infected with H. pylori strains having either the vacA s1m2 or s2m2 genotype. Many groups have found a correlation between vacuolating toxin activity and the pathogenicity of H. pylori, with the vacA s1m1 genotype having high toxin activity, s1m2 having intermediate activity, and s2m2 having low activity (3). Consistent with the findings in our study, in other western countries, infection with an H. pylori strain having the vacA s1m1 genotype is associated with more severe clinical disease (3, 33).
The original study from which these data were collected was a clinic-based, convenience sample of Alaskans with clinical symptoms requiring EGD evaluation. A limitation of our study, therefore, is that the comparison group for each clinical association was all persons without that diagnosis rather than strictly healthy persons. This could limit our ability to detect all clinical associations with the H. pylori cagA and vacA genotype. An additional limitation of our study is that over three-quarters of the participants were Alaska Native people, so the results presented here may not be generalizable outside Alaska. However, as rates of gastric cancer, PUD, and H. pylori infection are high in the Alaska Native people (14, 24, 35), it is important to specifically investigate organisms causing disease in this population. Finally, other H. pylori virulence factors, such as the vacA intermediate (i) region genotype and the blood group antigen binding adhesion (babA) genotype, may be equally, or more, important predictors of pathogenicity as the presence of the cagA gene and the vacA s and m region genotypes. It is also possible that the series of amino acids that make up the CagA protein, especially those of the EPIYA motifs, may be important predictors. It was beyond the scope of this project to test for these potential additional predictors of disease; however, we have plans to further analyze these samples as part of a future investigation of possible gastric cancer risk factors.
In conclusion, in this Alaskan population, where we have found high rates of H. pylori infection, gastric cancer, and PUD, we found that over 80% of H. pylori organisms carry the cagA gene, indicating the presence of the putative cag PAI virulence marker. In addition, almost half of the H. pylori strains contain the high-toxin-producing, and potentially more virulent, vacA s1m1 genotype. Infection with an H. pylori strain containing the cagA gene or the vacA s1m1 genotype was associated with a decreased incidence of esophagitis, supporting the hypothesis that infection with more virulent H. pylori organisms can protect persons from esophageal diseases. Similar to studies in other western parts of the world, infection with an H. pylori strain containing the vacA s1m1 genotype was also associated with more severe disease. Finally, two-thirds of the H. pylori organisms that had the less virulent vacA s2m2 genotype also contained the cagA gene, a novel finding for our population.
ACKNOWLEDGMENTS
We thank the following physicians who contributed patient data and biopsy specimens to this study: David Barrett, Richard Buchanan, Elaine Callahan, Mary Christian, Paul Davis, Bayne French, John Harvey, Mia Lee, Stephen Livingston, Patrick Martinez, Richard McGrath, David Powers, Gerry Sahagun, Charles Shannon, Thomas Shreves, Kevin Stange, James Stragand, Michael Swenson, Mark Thorndike, Steven Westby, and Frances Wilson. We thank the following nurses who recruited patients into the study: Catherine Dentinger, Marilyn Getty, Jim Gove, Cindy Hamlin, Helen Peters, and Susan Seidel. Finally, we thank Debra Parks for data management and Guillermo Perez-Perez for technical advice.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official positions of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Achtman M., et al. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459–470 [DOI] [PubMed] [Google Scholar]
- 2. Al Qabandi A., et al. 2005. Distribution of vacA and cagA genotypes of Helicobacter pylori in Kuwait. Acta Trop. 93:283–288 [DOI] [PubMed] [Google Scholar]
- 3. Atherton J. C., et al. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771–17777 [DOI] [PubMed] [Google Scholar]
- 4. Atherton J. C., et al. 1999. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J. Clin. Microbiol. 37:2979–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Azevedo N. F., Huntington J., Goodman K. J. 2009. The epidemiology of Helicobacter pylori and public health implications. Helicobacter 14(Suppl. 1):1–7 [DOI] [PubMed] [Google Scholar]
- 6. Backert S., et al. 2004. Functional analysis of the cag pathogenicity island in Helicobacter pylori isolates from patients with gastritis, peptic ulcer, and gastric cancer. Infect. Immun. 72:1043–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Backert S., Tegtmeyer N., Selbach M. 2010. The versatility of Helicobacter pylori CagA effector protein functions: the master key hypothesis. Helicobacter 15:163–176 [DOI] [PubMed] [Google Scholar]
- 8. Bruce M. G., Bruden D., McMahon B. J., Reasonover A., Morris J. 2007. The relationship between antimicrobial resistance and treatment outcome for Helicobacter pylori infections in Native and non-Native persons residing in Alaska. Helicobacter 12:450–451 [Google Scholar]
- 9. Bruce M. G., et al. 2006. Alaska sentinel surveillance for antimicrobial resistance in Helicobacter pylori isolates from Alaska native persons, 1999-2003. Helicobacter 11:581–588 [DOI] [PubMed] [Google Scholar]
- 10. Carothers J. J., et al. 2007. The relationship between previous fluoroquinolone use and levofloxacin resistance in Helicobacter pylori infection. Clin. Infect. Dis. 44:e5–e8 [DOI] [PubMed] [Google Scholar]
- 11. Chomvarin C., et al. 2008. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int. J. Infect. Dis. 12:30–36 [DOI] [PubMed] [Google Scholar]
- 12. Covacci A., et al. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. U. S. A. 90:5791–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cover T. L., Blaser M. J. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Demma L. J., et al. 2008. Epidemiology of hospitalizations associated with ulcers, gastric cancers and Helicobacter pylori infection among American Indian and Alaska Native persons. Am. J. Trop. Med. Hyg. 78:811–818 [PubMed] [Google Scholar]
- 15. Falush D., et al. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582–1585 [DOI] [PubMed] [Google Scholar]
- 16. Higashi H., et al. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. U. S. A. 99:14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hsu P. I., et al. 2002. Clinical presentation in relation to diversity within the Helicobacter pylori cag pathogenicity island. Am. J. Gastroenterol. 97:2231–2238 [DOI] [PubMed] [Google Scholar]
- 18. Huang J. Q., Sridhar S., Chen Y., Hunt R. H. 1998. Meta-analysis of the relationship between Helicobacter pylori seropositivity and gastric cancer. Gastroenterology 114:1169–1179 [DOI] [PubMed] [Google Scholar]
- 19. Kim S. Y., et al. 2001. Genotyping CagA, VacA subtype, IceA1, and BabA of Helicobacter pylori isolates from Korean patients, and their association with gastroduodenal diseases. J. Korean Med. Sci. 16:579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuipers E. J. 1999. Exploring the link between Helicobacter pylori and gastric cancer. Aliment. Pharmacol. Ther. 13(Suppl. 1):3–11 [DOI] [PubMed] [Google Scholar]
- 21. Kuipers E. J., Thijs J. C., Festen H. P. 1995. The prevalence of Helicobacter pylori in peptic ulcer disease. Aliment. Pharmacol. Ther. 9(Suppl. 2):59–69 [PubMed] [Google Scholar]
- 22. Kusters J. G., van Vliet A. H., Kuipers E. J. 2006. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19:449–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lai C. H., et al. 2002. High prevalence of cagA- and babA2-positive Helicobacter pylori clinical isolates in Taiwan. J. Clin. Microbiol. 40:3860–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lanier A. P., Maxwell J., McEnvoy T., Day G. E., Sandidge J. 2002. Alaska native cancer update 1988-2000 by sex, age service unit and year. Alaska Native Tribal Health Consortium, Anchorage, AK [Google Scholar]
- 25. Loffeld R. J., et al. 2000. Colonization with cagA-positive Helicobacter pylori strains inversely associated with reflux esophagitis and Barrett's esophagus. Digestion 62:95–99 [DOI] [PubMed] [Google Scholar]
- 26. Lu H. S., et al. 2008. Structural and functional diversity in the PAR1b/MARK2-binding region of Helicobacter pylori CagA. Cancer Sci. 99:2004–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lynn T. V., et al. 2007. Helicobacter pylori infection among non-Native educators in Alaska. Int. J. Circumpolar Health 66:135–143 [DOI] [PubMed] [Google Scholar]
- 28. Marshall B. J. 1995. The 1995 Albert Lasker Medical Research Award. Helicobacter pylori. The etiologic agent for peptic ulcer. JAMA 274:1064–1066 [DOI] [PubMed] [Google Scholar]
- 29. McMahon B. J., et al. 2006. Reinfection after successful eradication of Helicobacter pylori: a 2-year prospective study in Alaska Natives. Aliment. Pharmacol. Ther. 23:1215–1223 [DOI] [PubMed] [Google Scholar]
- 30. McMahon B. J., et al. 2003. The relationship among previous antimicrobial use, antimicrobial resistance, and treatment outcomes for Helicobacter pylori infections. Ann. Intern. Med. 139:463–469 [DOI] [PubMed] [Google Scholar]
- 31. Miehlke S., et al. 1996. Allelic variation in the cagA gene of Helicobacter pylori obtained from Korea compared to the United States. Am. J. Gastroenterol. 91:1322–1325 [PubMed] [Google Scholar]
- 32. Mukhopadhyay A. K., et al. 2000. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Navaglia F., et al. 1998. Helicobacter pylori cytotoxic genotype is associated with peptic ulcer and influences serology. Am. J. Gastroenterol. 93:227–230 [DOI] [PubMed] [Google Scholar]
- 34. Nomura A. M., Perez-Perez G. I., Lee J., Stemmermann G., Blaser M. J. 2002. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am. J. Epidemiol. 155:1054–1059 [DOI] [PubMed] [Google Scholar]
- 35. Parkinson A. J., et al. 2000. High prevalence of Helicobacter pylori in the Alaska native population and association with low serum ferritin levels in young adults. Clin. Diagn. Lab. Immunol. 7:885–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parsonnet J., Friedman G. D., Orentreich N., Vogelman H. 1997. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut 40:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pereira-Lima J. C., Marques D. L., Pereira-Lima L. F., Hornos A. P., Rota C. 2004. The role of cagA Helicobacter pylori strains in gastro- oesophageal reflux disease. Eur. J. Gastroenterol. Hepatol. 16:643–647 [DOI] [PubMed] [Google Scholar]
- 38. Sacco F., Bruce M. G., McMahon B. J., Bruden D. 2007. A prospective evaluation of 200 upper endoscopies performed in Alaska Native persons. Int. J. Circumpolar Health 66:144–152 [DOI] [PubMed] [Google Scholar]
- 39. Sgouros S. N., Bergele C. 2006. Clinical outcome of patients with Helicobacter pylori infection: the bug, the host, or the environment? Postgrad. Med. J. 82:338–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugimoto M., Yamaoka Y. 2009. The association of vacA genotype and Helicobacter pylori-related disease in Latin American and African populations. Clin. Microbiol. Infect. 15:835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Ende A., et al. 1998. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect. Immun. 66:1822–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van Doorn L. J., et al. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116:823–830 [DOI] [PubMed] [Google Scholar]
- 43. van Doorn L. J., et al. 1998. Expanding allelic diversity of Helicobacter pylori vacA. J. Clin. Microbiol. 36:2597–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Doorn L. J., et al. 1998. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 115:58–66 [DOI] [PubMed] [Google Scholar]
- 45. World Health Organization 1994. Infection with Helicobacter pylori. IARC Monogr. Eval. Carcinog. Risks Hum. 61:177–241 [PMC free article] [PubMed] [Google Scholar]
- 46. Yamaoka Y., et al. 1999. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J. Clin. Microbiol. 37:2274–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamaoka Y., et al. 2002. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 517:180–184 [DOI] [PubMed] [Google Scholar]
- 48. Yamaoka Y., et al. 2000. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol. Infect. 124:91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]