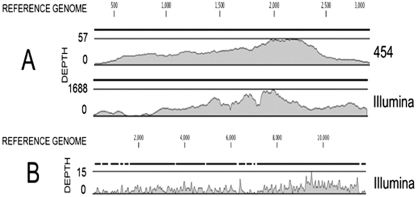
Fig. 1.
Distribution of the coverage resulting from de novo assembling or mapping on reference sequences of Roche-454 and Illumina reads. (A) Coverage of the anellovirus sequence by the contigs resulting from de novo assembling of Roche-454 and Illumina reads. Depth (y axis) corresponds to the number of reads representing a given nucleotide in the reconstructed sequence. The x axis corresponds to nucleotide positions in the whole genome. One contig from the Roche-454 covered 93.7% of the genome. Two contigs from the Illumina reads covered 92.6% of the genome. (B) Resequencing coverage of the genome of CVS strain of RABV from the hits directly mapped on the reference sequence. Note the difference of scale for the depth between the three graphs.