Abstract
The major goal of the present study was to investigate the potential use of a novel single nucleotide polymorphism (SNP) genotyping technology, called iPLEX Gold (Sequenom), for the simultaneous analysis of 16 SNPs that have been previously validated as useful for identification of Mycobacterium tuberculosis complex (MTBC) species and classification of MTBC isolates into distinct genetic lineages, known as principal genetic groups (PGGs) and SNP cluster groups (SCGs). In this context, we developed a 16-plex iPLEX assay based on an allele-specific-primer single-base-extension reaction using the iPLEX Gold kit (Sequenom), followed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis on the commercially available Sequenom MassARRAY platform. This assay was tested on a panel of 55 well-characterized MTBC strains that were also genotyped for the same loci using the previously reported SNaPshot assay, as well as 10 non-MTBC mycobacteria and 4 bacteria not belonging to the genus Mycobacterium. All MTBC samples were successfully analyzed with the iPLEX assay, which yielded clear allelic data for 99.9% of the SNPs (879 out of 880). No false-positive results were obtained with the negative controls. Compared to the SNaPshot assay, the newly developed 16-plex iPLEX assay produced fully concordant results that allowed reliable differentiation of MTBC species and recognition of lineages, thus demonstrating its potential value in diagnostic, epidemiological, and evolutionary applications. Compared to the SNaPshot approach, the implementation of the iPLEX technology could offer a higher throughput and could be a more flexible and cost-effective option for microbiology laboratories.
INTRODUCTION
The Mycobacterium tuberculosis complex (MTBC) is composed of causative agents of tuberculosis, a disease that remains a leading cause of human morbidity and mortality worldwide, with approximately 2 million deaths each year (World Health Organization, Tuberculosis Facts 2010 [http://www.who.int/tb/publications/factsheets/en/]). This complex comprises eight closely related bacterial species with distinct host tropisms, including the human pathogens M. tuberculosis, M. africanum, and M. canettii and the animal-adapted pathogens M. bovis, M. microti, M. caprae, and M. pinnipedii and the recently identified species M. mungi (1, 8, 12, 37). Although M. tuberculosis is the predominant causative agent of human tuberculosis, each member of this complex has been implicated in human infection, except M. mungi so far (8, 26). Moreover, two members, M. bovis, the causative agent of zoonotic bovine tuberculosis, and M. canettii, an unusual member responsible for rare tuberculosis cases almost always exposed to Africa, are naturally resistant to pyrazinamide, a first-line antituberculous drug (18, 34). Therefore, the rapid and reliable identification of MTBC isolates to the species level is of prime importance for timely selection of appropriate patient antibiotic treatment and also for epidemiological and public health considerations (36). Furthermore, various studies have recently identified distinct phylogenetic groupings within the human-adapted members of the MTBC (i.e., M. tuberculosis and M. africanum species), all of which are congruent (2, 5, 10, 14–16, 19, 20, 32). As shown in Table 1, these MTBC members are currently classified into six major phylogenetic lineages, two of which are composed of M. africanum strains. These six major lineages were first identified by analysis of genomic deletions or large sequence polymorphisms (LSPs), but they are highly congruent to the ones defined by single nucleotide polymorphisms (SNPs), such as principal genetic groups (PGGs) defined by Sreevatsan et al. (37) and SNP cluster groups (SCGs) defined by Filliol et al. (14). Some of the traditional groupings defined by the use of epidemiological tools (e.g., spoligotyping) also correlate with these lineages (9). These lineages are associated with particular geographical regions and show differences in their immunogenicities, virulence, and, possibly, drug susceptibilities (7, 16, 21, 32, 37). Thus, the recognition of these human-adapted MTBC lineages is useful to address evolutionary questions and can also provide information important for tuberculosis control (3).
Table 1.
Major phylogenetic lineages within human-adapted MTBC membersa
| Lineage | MTBC species | Presence/absence of TbD1 | Gagneux's nomenclature (LSP based) | PGG (SNP based) | SCG (SNP based) | Spoligotype-defined families |
|---|---|---|---|---|---|---|
| 1 | M. tuberculosis | Intact | Indo-Oceanic lineage | 1b | 1 | EAI |
| 2 | M. tuberculosis | Deleted | East Asian lineage | 1b | 2 | Beijing |
| 3 | M. tuberculosis | Deleted | East African-Indian lineage | 1b | 3a | CAS |
| 4 | M. tuberculosis | Deleted | Euro-American lineage | 2, 3 | 3b, 3c, 4, 5, 6a, 6b | H, LAM, X, T, S, others |
| 5 | M. africanum | Intact | West African lineage I | 1b | Not described | AFRI2 |
| 6 | M. africanum | Intact | West African lineage II | 1a | Not described | AFRI1 |
TbD1, M. tuberculosis-specific deletion region 1 described by Brosch et al. (8); PGGs are as defined by Sreevatsan et al. (37); SCG subgroups are as defined by Filliol et al. (14) and Alland et al. (2); EAI, East African-Indian family; CAS, Central Asian family; H, Haarlem family; LAM, Latin American-Mediterranean family.
In the last decade, a number of nucleic acid-based amplification methods have been developed for the differentiation of MTBC species in an attempt to replace the time-consuming analysis of the phenotypic and biochemical characteristics of the bacteria after culture, which has long been the “gold standard” (4, 17, 23, 25, 27, 28, 30, 38). All these PCR-based methods, including the commercial GenoType MTBC line-probe assay (Hain Lifescience), are based on the analysis of species-specific polymorphisms, which are generally SNPs and/or LSPs (or regions of difference [RDs], e.g., genomic deletions). The recently described exact tandem repeat D (ETR-D) sequencing method is an alternative approach that enables MTBC species identification, thanks to (i) variable numbers (1 to 7 copies) of the tandem repeat, (ii) six specific SNPs, and (iii) two deletions/insertions (13). However, none of these methods allow the definitive identification of MTBC isolates to the species level and their simultaneous classification into lineages, although LSPs and SNPs are also ideal markers for defining MTBC phylogenetic groupings (11, 16).
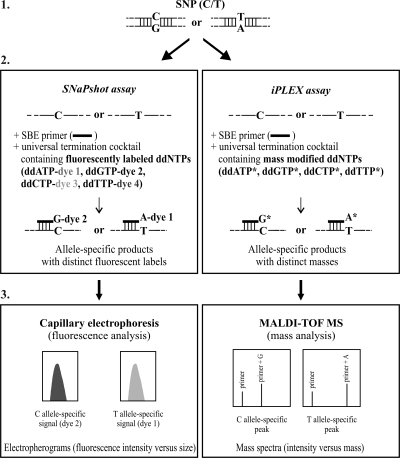
In this context, we recently reported the development of an innovative two-step strategy targeting 16 species- and lineage-specific MTBC SNPs using two 8-plex assays based on the SNaPshot technology (Applied Biosystems [AB], Foster City, CA) (6). As illustrated in Fig. 1, this SNP genotyping technology combines allele-specific-primer single-base-extension (SBE) biochemistry using fluorescent terminators with capillary electrophoresis detection. The first 8-plex SNaPshot assay enables the recognition of the three PGGs defined by Sreevatsan et al. (37) and the reliable identification of MTBC members (except PGG-1b M. tuberculosis and PGG-1b M. africanum and M. mungi), while the second 8-plex SNaPshot assay enables the further classification of M. tuberculosis isolates into 1 of the 6 SCGs and 5 subgroups defined by Filliol et al. (14). As supported by our results (6), a unique feature of this two-step strategy is that it allows the simultaneous identification of MTBC species and lineages using the combination of the two 8-plex assays.
Fig. 1.
Principle of SNP analysis by using two independent SNP genotyping technologies: the SNaPshot technology (Applied Biosystems) and the iPLEX technology (Sequenom). These genotyping methods comprise three main steps, namely, PCR amplification of the DNA region containing the polymorphic site (step 1), an allelic discrimination reaction consisting of an SBE reaction (step 2), and detection of allele-specific products (step 3).
In recent years, many SNP genotyping technologies, including fully integrated commercial solutions, have become available (29, 31, 35). On the basis of different biochemistries and detection platforms, each offers a unique combination of sample throughput, multiplexing capability, and cost. In the present study, we investigated the potential use of a novel SNP genotyping technology based on the commercially available Sequenom Inc. (San Diego, CA) MassARRAY platform for the simultaneous analysis of the 16 SNPs that we previously selected for the identification of MTBC species and lineages by the SNaPshot approach (6). Like the SNaPshot technology, the recent iPLEX Gold technology (Sequenom) relies upon a primer SBE reaction for allelic discrimination, but it uses mass-modified terminators that are nonfluorescent, and SBE products are detected by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), as illustrated in Fig. 1. Thus, the iPLEX Gold technology combines the benefits of robust primer SBE biochemistry with the sensitivity, rapidity, and accuracy of MALDI-TOF MS detection. The newly developed 16-plex iPLEX assay was evaluated in comparison to a modified version of the previously developed SNaPshot approach, allowing the simultaneous analysis of the 16 SNPs in a single 16-plex SNaPshot assay.
MATERIALS AND METHODS
Bacterial samples.
This study is based on the collection of MTBC DNA samples analyzed in our previous work (6). A total of 55 MTBC DNA samples from reference strains M. tuberculosis H37Rv ATCC 27294, M. bovis CIP 102426, and M. bovis BCG CIP 105226 and clinical isolates of M. tuberculosis (n = 34), M. bovis (n = 6), M. bovis BCG (n = 4), M. africanum (n = 4), M. canettii (n = 1), M. caprae (n = 1), M. microti (n = 1), and M. pinnipedii (n = 1) were used in the present study. These isolates were identified to the species level by phenotypic and biochemical characterization methods and/or by use of a gene probe assay and, for some of them, by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) analysis, as previously described (6). In total, 37 of these samples (M. tuberculosis H37Rv, M. bovis CIP 102426, one clinical M. africanum isolate, and all clinical M. tuberculosis isolates) had previously been successfully genotyped for the 16 SNP loci targeted in this study, and all the other ones has been successfully genotyped for 8 of these SNP loci. DNA samples from 10 mycobacteria other than those belonging to the MTBC, including M. fortuitum (n = 1), M. kansasii (n = 2), M. abscessus (n = 1), M. avium (n = 3), M. chelonae (n = 2), and M. gordonae (n = 1), and 4 bacteria that do not belong to the genus Mycobacterium (Nocardia nova, Corynebacterium amycolatum, Staphylococcus aureus, and Escherichia coli) were also used in this study as negative controls.
SNP panel.
The assays developed in this study were designed to target a panel of 16 MTBC phylogenetically relevant SNPs previously selected on the basis of their species or lineage specificity (6). General information about these SNPs is given in Table 2. It must be noted here that for technical reasons, the SNP at position 2460628 (i.e., position 2460626 in the work of Filliol et al. [14]) targeted by the 16-plex SNaPshot assay was replaced by SNP at position 144390 for the 16-plex iPLEX assay (numbering according to the M. tuberculosis H37Rv genome, GenBank accession no. NC_000962.2). These two polymorphisms are phylogenetically coincident and mark SCG-4 M. tuberculosis strains (2, 14).
Table 2.
General information on SNPs analyzed in this study
| SNP name | H37Rv |
Polymorphism | Usefulness | Reference(s) | |
|---|---|---|---|---|---|
| Positionb | Alleleb | ||||
| katG463 | 2154724 | C | C/A | PGG and SCG assignation | 37, 2 |
| gyrA95 | 7585 | G | G/C | PGG assignation | 37 |
| katG203 | 2155503 | G | G/A | Segregation of PGG-1a from PGG-1b isolates | 14a |
| hsp65631 | 529006 | C | C/T | Differentiation of M. canettii from other MTBC species | 18 |
| 16S rRNA1249 | 1473094 | T | T/C | Differentiation of M. pinnipedii from other MTBC species | 23a |
| gyrB(675) | 5671 | C | C/T | Differentiation of M. microti from other MTBC species | 25, 27 |
| gyrB(756) | 5752 | G | G/A | Differentiation of M. caprae and M. bovis from other MTBC species | 25, 27 |
| gyrB(1410) | 6406 | C | C/T | Differentiation of M. bovis from other MTBC species | 25, 27 |
| 1977 | 1977 | A | A/G | SCG assignation | 2 |
| 3352929 | 3352932 | C | C/G | SCG assignation | 14 |
| 74092 | 74092 | C | C/T | SCG assignation | 2 |
| 105139 | 105139 | C | C/A | SCG assignation | 2 |
| 144390a | 144390 | G | G/A | SCG assignation | 2 |
| 232574 | 232574 | G | G/T | SCG assignation | 2 |
| 311613 | 311613 | G | G/T | SCG assignation | 2 |
| 913274 | 913274 | C | C/G | SCG assignation | 2 |
| 2460626a | 2460628 | C | C/A | SCG assignation | 14 |
The two SNPs 144390 and 2460626 are phylogenetically coincident since the A in both cases is characteristic of M. tuberculosis SCG-4 lineage.
Positions and alleles are relative to the plus strand on the M. tuberculosis H37Rv genome sequence, GenBank accession no. NC_000962.2.
Modified 16-plex SNaPshot assay.
In order to better compare the SNaPshot and the iPLEX technologies, we combined the two 8-plex SNaPshot assays described by Bouakaze et al. (6) into a single 16-plex SNaPshot assay allowing the simultaneous analysis of the 16 SNPs of interest. PCR and SBE primers previously designed were first tested without modification in a single 16-plex SNaPshot assay with 1 ng DNA of M. tuberculosis H37Rv as template as previously described, except that capillary electrophoresis was performed on an ABI 3500 genetic analyzer (AB, Foster City, CA), and automated allele calling was done using GeneMapper (version 4) software (AB) (6). However, to enable the automated allele calling for all 16 SNPs, we had to modify the sequences of three SBE primers by either increasing or reducing the size of their 5′ tail so that the peak for each extension product was easily distinguishable on the electropherograms. The sizes of the SBE primers for SNP loci 2460628 and 105139 were increased to 65 nucleotides (nt) instead of 55 nt and 52 nt instead of 50 nt, respectively, while the SBE primer size for SNP locus gyrB(675) was reduced from 64 nt to 52 nt. All 55 MTBC samples were finally analyzed again for these 16 loci using the 16-plex SNaPshot assay with the newly ordered primers.
Novel 16-plex iPLEX assay.
PCR and SBE primers for each SNP investigated were designed using the MassARRAY design software, version 4.0 (Sequenom Inc., San Diego, CA), with the exception of primers for SNP loci hsp65631 and 144390, which were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/). The optimal amplicon size was set to 80 to 120 bp. A 10-mer tag (5′-ACGTTGGATG-3′) was added to the 5′ end of each PCR primer to avoid confusion in the mass spectrum, and SBE primers were 5′ tailed with nonhomologous sequences varying in length to create large enough mass differences between the different SBE products to be detected by MALDI-TOF MS. PCR and SBE primer sequences are shown in Table 3.
Table 3.
PCR and SBE primers used in this study for the 16-plex iPLEX assay
| SNP locus | Second PCR primera | First PCR primera | Amplicon length (bp) | UEP directionb | SBE unextended primer (UEP)a |
|---|---|---|---|---|---|
| 913274 | acgttggatgATGGTGTACTGCTGCTTGAG | acgttggatgACGTGTTGCTGATGGACGAG | 117 | F | GCTGGACCCAATCTC |
| 74092 | acgttggatgAAAGAGCGCTACGCCAGATG | acgttggatgGTACTCCTTCACCGCCTTG | 116 | F | CGAATTGCCTTGGCG |
| 16S rRNA1249 | acgttggatgACCGGCTTTTAAGGATTCGC | acgttggatgTTATGTCCAGGGCTTCACAC | 105 | F | GCCGGTACAAAGGGC |
| gyrB(756) | acgttggatgGTGGTTTCGAAAACAGCGGG | acgttggatgATGAGAAGTCGGAACCCCTG | 120 | F | gGACGGGGTCAACGGT |
| gyrA95 | acgttggatgTCCACCAGCGGGTAGCGCA | acgttggatgAGACCATGGGCAACTACCAC | 117 | F | CGCGTCGATCTACGACA |
| 311613 | acgttggatgACTTGCTACGCGTCCTACC | acgttggatgGATGTTCTTGTCGCCCAGAG | 115 | F | GCCGTTCGTAGCCGCAG |
| hsp65631 | acgttggatgGTCTCAAACGCGGCATCGAAAAG | acgttggatgAATCTGCTCCTTGGTCTCGACCTC | 109 | R | aGTCTCGACCTCCTTGGC |
| katG203 | acgttggatgTTACCGCTGTAACGCTCATC | acgttggatgGAGCCCGATGAGGTCTATTG | 85 | R | ggaGGGCAAGGAAGCCAC |
| 3352929 | acgttggatgACGGTCCGCAACCACAATC | acgttggatgGTCGCAAGCATCTGACATTG | 93 | R | TGACATTGGTGCACAAAAC |
| katG463 | acgttggatgGTCGAAACTAGCTGTGAGAC | acgttggatgGAGATTGCCAGCCTTAAGAG | 85 | R | cctCCTTAAGAGCCAGATCC |
| gyrB(1410) | acgttggatgACAGCCTTGTTCACAACGAC | acgttggatgCGAGGTCAAATCGTTTGTGC | 119 | F | gTTGTGCAGAAGGTCTGTAA |
| gyrB(675) | acgttggatgTCCGACTTCTCATAAACCTG | acgttggatgTGGTTAACGCGCTATCCACC | 102 | F | ggggTCAAGCGCGACGGGTA |
| 232574 | acgttggatgACCCCAGTGCCTTCAGAAAG | acgttggatgAAGATCTTCTACTACGGCGG | 102 | F | gctGACAGGGCAATCACCTCG |
| 105139 | acgttggatgTTCGAATCATAACGTCGGGC | acgttggatgCTGGGTAAATCCCTTGTGTC | 101 | R | cccatTTTCACGGTTATCAGCG |
| 1977 | acgttggatgTAGCGTGGGGACTGCCAAC | acgttggatgTACGGTTGTTGTTCGACTGC | 102 | F | tggctGGTCACGCGTCATGGGC |
| 144390 | acgttggatgTGGGTTCTGCCCTGTCGTG | acgttggatgATCACCGAGCAACGCGTCC | 88 | F | cggttcGGTGGTGCTGATTGACGC |
The lowercase letters in the primer sequences are 5′-end tags (PCR primers) or nonhomologous sequences (SBE primers) that were added to increase the molecular weights of these primers so that they do not interfere with expected products in the mass spectra.
F, forward; R, reverse.
The genotyping analysis was performed as recommended by the manufacturer with reagents included in the iPLEX Gold SNP genotyping kit (Sequenom) and the software and equipment provided with the MassARRAY platform (Sequenom). The 16 target sequences were simultaneously amplified from a 5-μl final PCR volume composed of 1× PCR buffer, 2 mM MgCl2, 500 μM deoxynucleoside triphosphates (dNTPs), 0.1 μM each PCR primer, 0.5 U of HotStarTaq enzyme, and 3.4 μl bacterial DNA extract. The thermal cycling conditions consisted of a first denaturation step at 95°C for 2 min, followed by 45 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min, with a final extension step at 72°C for 5 min. To neutralize unincorporated dNTPs, PCR products were treated with 0.5 U shrimp alkaline phosphatase by incubation at 37°C for 40 min, followed by enzyme inactivation by heating at 85°C for 5 min. By adding 2 μl of an iPLEX Gold extension reaction cocktail to the purified PCR products, the 16-plex SBE reaction was carried out in a final volume of 9 μl containing 0.222× iPLEX buffer, 1× iPLEX termination mix, 1× iPLEX enzyme, and the SBE primer mix that contained the 16 SBE primers divided into 4 groups from low to high masses (each group was composed of 4 primers). In the final SBE reaction, the concentration of the low-mass primers mix was 7 μM, the concentrations of the two medium-mass primer mixes were 9.3 μM and 11.6 μM, and the concentration of high-mass primers was 14 μM. The iPLEX extension reaction was performed under the following thermal conditions: an initial denaturation step at 94°C for 30 s, followed by 40 cycles of a denaturation step at 94°C for 5 s, 5 cycles of annealing at 52°C for 5 s and extension at 80°C for 5 s, and finally, a final extension step at 72°C for 3 min. After desalting of the products by using SpectroCLEAN resin following the manufacturer's protocol, cleaned extension products were dispensed onto a 96 SpectroCHIP array using an RS1000 Nanodispenser, and finally, the array was introduced into a MassARRAY Compact 96 mass spectrometer. Spectra were acquired using SpectroAcquire software, and data analysis, including automated allele calling, was done using MassARRAY Typer software, version 4.0.5. At least two replicate experiments were performed for each sample.
DNA sequencing.
Markers that failed to be detected with the 16-plex SNaPshot assay and/or 16-plex iPLEX assay were amplified in singleplex reactions using SNaPshot primer pairs as previously described (6). After visual assessment of PCR products by agarose gel electrophoresis, standard sequencing was performed in both directions using a BigDye Terminator (version 3.1) cycle sequencing kit (AB) according to the manufacturer's recommendations. Capillary electrophoresis was performed on an ABI 3500 genetic analyzer (AB), and the resulting sequences were assembled and edited using the software Sequencher, version 4.7 (Gene Codes Corp., Ann Arbor, MI).
RESULTS
The 16 SNPs that were targeted by the previously described two 8-plex SNaPshot assays for identification of M. tuberculosis complex species and lineages (6) were successfully analyzed by using the modified 16-plex SNaPshot assay described in this study. The 55 MTBC samples tested with this single 16-plex SNaPshot assay yielded allelic data fully concordant with those obtained previously with the two 8-plex SNaPshot assays. The M. canettii sample still unexpectedly failed to amplify 3 loci (gyrA95, 1977, and 105139). Thus, 877 alleles were assigned over the 880 alleles that were expected, resulting in an allele call rate of 99.7%. Negative controls analyzed with the 16-plex SNaPshot assay failed to amplify all SNP loci, except sometimes for some loci (hsp65631, 16S rRNA1249, and 232574), as also previously observed with the two 8-plex SNaPshot assays (6).
For comparison, the new 16-plex iPLEX assay was tested on the same set of 55 MTBC samples. It resulted in 879 allele calls, yielding an allele call rate of 99.9%. The alleles that were duplicated between the 16-plex iPLEX and SNaPshot assays were 100% concordant. The only one allele that was not assigned, even after a manual check, corresponded to that of the M. canettii sample for the SNP at position 105139. The full set of genotyping results obtained by the iPLEX technology for the 55 MTBC samples analyzed in this study, as well as the PGGs and SCGs inferred from these genotypes, is reported in Table 4. All MTBC species tested were clearly differentiated in at least one locus, except for one M. africanum and two M. tuberculosis strains that displayed identical allelic combinations and that were clustered into the same group (PGG-1b/SCG-1). Therefore, the 16-plex iPLEX assay developed in this study is suitable for identification of MTBC species, except PGG-1b M. africanum and PGG-1b M. tuberculosis and M. mungi. This assay also has the potential to identify the main MTBC lineages, since the genotype data obtained for the lineage-specific markers enabled us to unambiguously classify all 55 MTBC samples analyzed in this study into one PGG and one SCG and no phylogenetic inconsistencies were observed. It must be noted that unexpected peaks were seen for some SNPs in negative-control samples, some of them being systematically observed even in water blanks, but this never interfered with data interpretation.
Table 4.
SNP genotyping results of the 16-plex iPLEX assay on the 55 MTBC samples tested in this study
| MTBC sample (n = 55) | PGGa | SCGb | Genotype |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsp65631 | katG463* | gyrA95 | katG203 | 16S rRNA1249 | gyrB(675) | gyrB(756) | gyrB(1410) | 1977* | 3352929* | 74092* | 105139* | 144390* | 232574* | 311613* | 913274* | |||
| Clinical M. canettii (n = 1) | 1b | 1 | T | A | C | G | T | C | G | C | G | G | C | – | G | G | T | G |
| Clinical M. tuberculosis (n = 2) and M. africanum (n = 1) | 1b | 1 | C | A | C | G | T | C | G | C | G | G | C | C | G | G | T | G |
| Clinical M. tuberculosis (n = 3) | 1b | 2 | C | A | C | G | T | C | G | C | G | G | C | A | G | G | T | C |
| Clinical M. tuberculosis (n = 2) | 1b | 3a | C | A | C | G | T | C | G | C | G | G | C | C | G | G | T | C |
| Clinical M. tuberculosis (n = 9) | 2 | 3b | C | C | C | G | T | C | G | C | G | G | C | C | G | G | T | C |
| Clinical M. tuberculosis (n = 13) | 2 | 5 | C | C | C | G | T | C | G | C | G | C | C | C | G | G | T | C |
| Clinical M. tuberculosis (n = 5) | 3 | 6a | C | C | G | G | T | C | G | C | A | C | C | C | G | G | T | C |
| M. tuberculosis H37Rv (n = 1) | 3 | 6b | C | C | G | G | T | C | G | C | A | C | C | C | G | G | G | C |
| Clinical and reference M. bovis (n = 7) and M. bovis BCG (n = 5) | 1a | 7 | C | A | C | A | T | C | A | T | G | G | T | C | G | G | T | G |
| Clinical M. caprae (n = 1) | 1a | 7 | C | A | C | A | T | C | A | C | G | G | T | C | G | G | T | G |
| Clinical M. microti (n = 1) | 1a | 1 | C | A | C | A | T | T | G | C | G | G | C | C | G | G | T | G |
| Clinical M. pinnipedii (n = 1) | 1a | 1 | C | A | C | A | C | C | G | C | G | G | C | C | G | G | T | G |
| Clinical M. africanum (n = 3) | 1a | 1 | C | A | C | A | T | C | G | C | G | G | C | C | G | G | T | G |
MTBC isolates were assigned to one of the three PGGs delineated by Sreevatsan et al. (37) on the basis of allelic combination at 3 loci (katG463, gyrA95, and katG203).
MTBC isolates were assigned to one of the seven SCGs delineated by Filliol et al. (14) on the basis of allelic combination at 9 loci (polymorphisms marked with an asterisk).
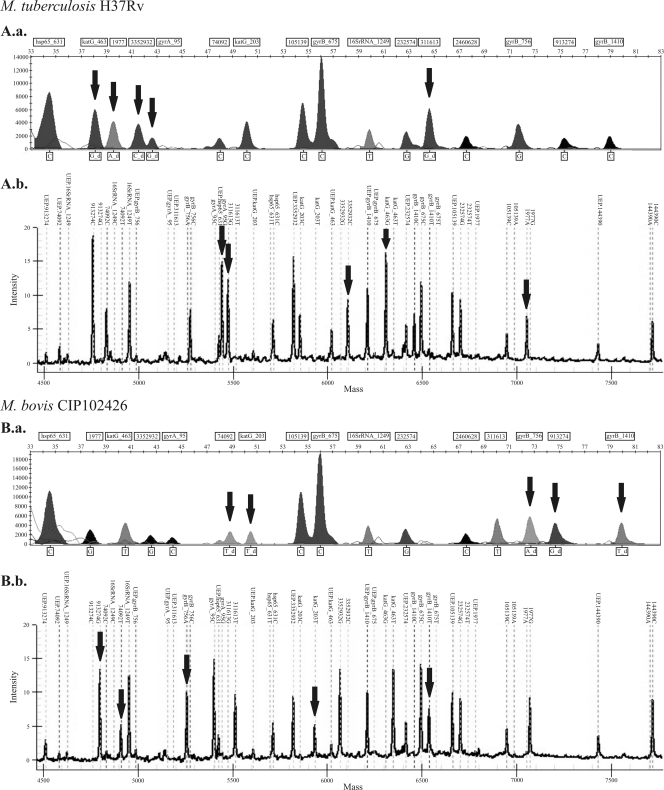
Figure 2 shows examples of electropherograms and mass spectra obtained for the reference strains M. tuberculosis H37Rv and M. bovis CIP 102426 using the 16-plex SNaPshot assay and the 16-plex iPLEX assay, respectively. As illustrated, these two species yielded distinct electropherograms and mass spectra, and for each species, the genotyping results obtained with the two different SNP genotyping assays were concordant with each other and with data in the literature (14, 37). Indeed, the M. tuberculosis H37Rv sample was always found to have the combinations of polymorphisms characteristic of PGG-3 (i.e., CTG → CGG at katG codon 463 and ACC → AGC at gyrA codon 95) and SCG-6b (1977G → A, 3352932G → C, and 311613T → G). In contrast, the M. bovis sample possessed the polymorphisms associated with PGG-1a (i.e., ACC → ACT at katG codon 203) and SCG-7 (74092C → T and 913274C → G), as well as the mutated alleles for two gyrB gene SNPs, gyrB (756G → A) and the M. bovis-specific gyrB polymorphism (1410C → T).
Fig. 2.
M. tuberculosis H37Rv and M. bovis CIP102426 electropherograms and mass spectra obtained using the 16-plex SNaPshot assay and 16-plex iPLEX assay, respectively. Electropherograms (A.a. and B.a.) were generated with GeneMapper (version 4) software (AB) and show the relative fluorescence units (RFUs) versus the measured size (in nucleotides) of the SBE products relative to the GeneScan-120 LIZ internal size standard (AB). Mass spectra (A.b. and B.b.) were generated using MassARRAY Typer (version 4.0.5) software (Sequenom) and show the relative intensity versus the mass of the analytes. Mutated alleles are indicated by arrows.
The DNA regions surrounding the three loci that failed to be analyzed from the M. canettii sample with the SNaPshot assay and/or the iPLEX assay (i.e., 105139, 1977, and gyrA95) were further investigated by DNA sequencing. The sequencing results revealed that the M. canettii sample used in this study showed sequence variations in the SNaPshot SBE primer binding sites for these three loci and also in the iPLEX SBE primer binding site for the 105139 locus. Thus, the primer extension failures were very likely caused by mismatches between the SBE primers and their targets, preventing the extension reactions. Although we failed to detect these 3 loci from the M. canettii DNA sample, the two assays described in this study easily differentiated M. canettii from the other species thanks to the C-to-T substitution at locus hsp65631, which is a polymorphism previously reported to be specific for the M. canettii species (17).
DISCUSSION
In a previous study, we reported the development of two complementary 8-plex SNaPshot assays for the analysis of 16 SNPs that enable (i) the identification of MTBC members (except PGG-1b M. africanum and PGG-1b M. tuberculosis and M. mungi), (ii) the recognition of PGG lineages as defined by Sreevatsan et al. (37), and (iii) the classification of M. tuberculosis isolates into one of the six SCG lineages defined by Filliol et al. (14). The present study confirms that the simultaneous analysis of these 16 SNPs can be achieved in an equally efficient and reliable manner using a single 16-plex SNaPshot assay according to the protocol described in our previous article and modified as described herein. Thus, our results support the fact that SNaPshot assays can be readily multiplexed to a level higher than that suggested by the manufacturer, which recommends a limit of 10 SNPs per assay. As multiplexing is an efficient way to reduce costs and increase throughput, the use of this 16-plex SNaPshot assay is a cost-efficient and time-saving alternative option for laboratories that are interested in both identification of MTBC species and recognition of PGGs and SCGs by using a capillary electrophoresis platform.
This study also describes the development of a new 16-plex iPLEX assay for genotyping of these species- and lineage-specific SNPs using the commercially available Sequenom MassARRAY MALDI-TOF MS platform. This 16-plex iPLEX assay generated a high SNP call rate and showed a high degree of reproducibility. A perfect concordance was observed compared to the data generated by the 16-plex SNaPshot assay. Therefore, the newly developed 16-plex iPLEX assay can also be used for reliable identification of MTBC species and recognition of PGGs and SCGs from cultured MTBC strains.
The SNaPshot and iPLEX assays described in this study are both effective and easy to use and produce data that are easy to interpret since alleles are automatically called by ad hoc analysis softwares. Nevertheless, each assay presents specific advantages and disadvantages since the assays are based on different SNP genotyping technologies. The major advantage of the SNaPshot-based assay is that it can easily be introduced in a laboratory having access to an automated sequencer, which is equipment now commonly found in many microbiology laboratories for other common applications, such as classical DNA sequencing and MIRU-VNTR typing. However, this approach requires the use of fluorescently labeled terminators, which is not the case for the iPLEX assay. The iPLEX assay offers many additional benefits with respect to the SNaPshot assay. For instance, an analysis by MALDI-TOF MS is much faster than an analysis by capillary electrophoresis, requiring a few seconds for the former one and up to several minutes for the latter one. In addition, the iPLEX assay is suitable for high-throughput analysis, as either 96 or 384 samples can be analyzed on the same chip, depending on the MassARRAY platform used. Furthermore, a recent study has shown that spoligotyping of the MTBC members by analysis of the 43 spacers found in the direct repeat region can be done on the MassARRAY platform (22). The analysis of these markers, which is currently routinely performed by a reverse line blot hybridization assay, is very useful for the molecular characterization/genotyping of MTBC strains for epidemiological purposes (9, 24). Thus, the MassARRAY platform is also able to provide valuable information for molecular typing of MTBC strains. Although the major drawback of the iPLEX assay lies in the requirement for specific equipment, investment in a MassARRAY platform can be very appealing for microbiology centers with medium- to high-throughput activities, as previously noticed (33).
To conclude, this study demonstrated that the iPLEX technology with the MassARRAY platform can be used for accurate genotyping of 16 SNPs that enable simultaneous differentiation of MTBC species and characterization of the main phylogenetic lineages. Compared to the 16-plex SNaPshot assay, the 16-plex iPLEX assay could offer a higher throughput and a more flexible and cost-effective option for microbiology laboratories. Nevertheless, this study represents only an initial evaluation of the use of this MALDI-TOF MS-based assay for identification of MTBC species and lineages and requires further evaluations with larger collections of MTBC samples.
ACKNOWLEDGMENTS
We particularly thank Susanne Müller at Sequenom (Hamburg, Germany) for her valuable technical help during the development of the iPLEX assay.
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Alexander K. A., et al. 2010. Novel Mycobacterium tuberculosis complex pathogen, M. mungi. Emerg. Infect. Dis. 16: 1296–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alland D., et al. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 45: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allix-Beguec C., Harmsen D., Weniger T., Supply P., Niemann S. 2008. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 46: 2692–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnold C., et al. 2005. Single-nucleotide polymorphism-based differentiation and drug resistance detection in Mycobacterium tuberculosis from isolates or directly from sputum. Clin. Microbiol. Infect. 11: 122–130 [DOI] [PubMed] [Google Scholar]
- 5. Baker L., Brown T., Maiden M. C., Drobniewski F. 2004. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg. Infect. Dis. 10: 1568–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouakaze C., et al. 2010. Identification and genotyping of Mycobacterium tuberculosis complex species by use of a SNaPshot minisequencing-based assay. J. Clin. Microbiol. 48: 1758–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brimacombe M., Hazbon M., Motiwala A. S., Alland D. 2007. Antibiotic resistance and single-nucleotide polymorphism cluster grouping type in a multinational sample of resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 51: 4157–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brosch R., et al. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99: 3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brudey K., et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Comas I., et al. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Comas I., Homolka S., Niemann S., Gagneux S. 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4: e7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cousins D. V., et al. 2003. Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipedii sp. nov. Int. J. Syst. Evol. Microbiol. 53: 1305–1314 [DOI] [PubMed] [Google Scholar]
- 13. Djelouadji Z., Raoult D., Daffe M., Drancourt M. 2008. A single-step sequencing method for the identification of Mycobacterium tuberculosis complex species. PLoS Negl. Trop. Dis. 2: e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Filliol I., et al. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188: 759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a. Frothingham R., et al. 1999. Phenotypic and genotypic characterization of Mycobacterium africanum isolates from West Africa. J. Clin. Microbiol. 37: 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gagneux S., et al. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103: 2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gagneux S., Small P. M. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7: 328–337 [DOI] [PubMed] [Google Scholar]
- 17. Goh K. S., et al. 2006. Study of the gyrB gene polymorphism as a tool to differentiate among Mycobacterium tuberculosis complex subspecies further underlines the older evolutionary age of ‘Mycobacterium canettii.’ Mol. Cell. Probes 20: 182–190 [DOI] [PubMed] [Google Scholar]
- 18. Goh K. S., Legrand E., Sola C., Rastogi N. 2001. Rapid differentiation of “Mycobacterium canettii” from other Mycobacterium tuberculosis complex organisms by PCR-restriction analysis of the hsp65 gene. J. Clin. Microbiol. 39: 3705–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gutacker M. M., et al. 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J. Infect. Dis. 193: 121–128 [DOI] [PubMed] [Google Scholar]
- 20. Hershberg R., et al. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6: e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirsh A. E., Tsolaki A. G., DeRiemer K., Feldman M. W., Small P. M. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. U. S. A. 101: 4871–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Honisch C., et al. 2010. Replacing reverse line blot hybridization spoligotyping of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 48: 1520–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huard R. C., Lazzarini L. C., Butler W. R., van Soolingen D., Ho J. L. 2003. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 41: 1637–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a. Huard R. C., et al. 2006. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J. Bacteriol. 188: 4271–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamerbeek J., et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35: 907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kasai H., Ezaki T., Harayama S. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38: 301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiers A., Klarenbeek A., Mendelts B., Van Soolingen D., Koeter G. 2008. Transmission of Mycobacterium pinnipedii to humans in a zoo with marine mammals. Int. J. Tuberc. Lung Dis. 12: 1469–1473 [PubMed] [Google Scholar]
- 27. Niemann S., Harmsen D., Rusch-Gerdes S., Richter E. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38: 3231–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parsons L. M., et al. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40: 2339–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perkel J. 2008. SNP genotyping: six technologies that keyed a revolution. Nat. Methods 5: 447–453 [Google Scholar]
- 30. Pinsky B. A., Banaei N. 2008. Multiplex real-time PCR assay for rapid identification of Mycobacterium tuberculosis complex members to the species level. J. Clin. Microbiol. 46: 2241–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ragoussis J. 2009. Genotyping technologies for genetic research. Annu. Rev. Genomics Hum. Genet. 10: 117–133 [DOI] [PubMed] [Google Scholar]
- 32. Reed M. B., et al. 2009. Major Mycobacterium tuberculosis lineages associate with patient country of origin. J. Clin. Microbiol. 47: 1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sauer S., Kliem M. 2010. Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 8: 74–82 [DOI] [PubMed] [Google Scholar]
- 34. Scorpio A., Zhang Y. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2: 662–667 [DOI] [PubMed] [Google Scholar]
- 35. Sobrino B., Brion M., Carracedo A. 2005. SNPs in forensic genetics: a review on SNP typing methodologies. Forensic Sci. Int. 154: 181–194 [DOI] [PubMed] [Google Scholar]
- 36. Somoskovi A., et al. 2009. “Mycobacterium canettii” isolated from a human immunodeficiency virus-positive patient: first case recognized in the United States. J. Clin. Microbiol. 47: 255–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sreevatsan S., et al. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. U. S. A. 94: 9869–9874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Warren R. M., et al. 2006. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int. J. Tuberc. Lung Dis. 10: 818–822 [PubMed] [Google Scholar]