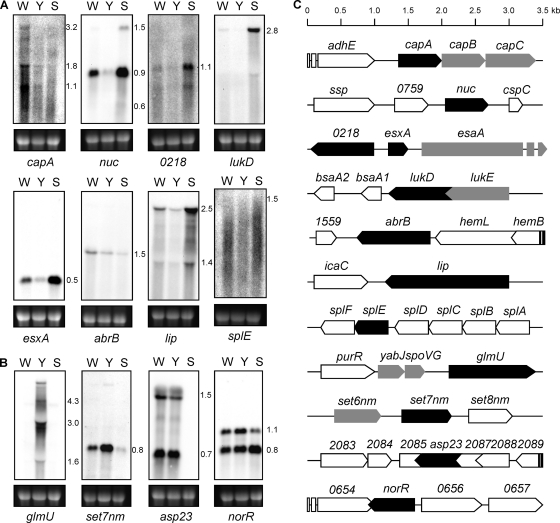
Fig. 1.
Northern blot analysis of yabJ-spoVG-dependent genes. Northern blots of total RNA from S. aureus Newman (W), ΔyabJ-spoVG mutant SM148 (Y), and ΔsigB mutant MS64 (S) for selected genes that were found to be down- (A) or upregulated (B) in the microarray in SM148. Transcript masses are indicated in kb. The ethidium bromide-stained 16S rRNA pattern is shown as an indication for the RNA loading. (C) Locus maps of ORFs selected for Northern blots (black). ORFs appearing in the microarray are highlighted in gray. adhE, encoding iron-containing alcohol dehydrogenase; ssp, encoding extracellular matrix and plasma binding protein; cspC, encoding cold-shock protein CSD family protein; bsaA1/2, encoding lantibiotic precursors; hemL, encoding glutamate-1-semialdehyde-2,1-aminomutase; hemB, encoding delta-aminolevulinic acid dehydratase; icaC, encoding intercellular adhesion protein IcaC; splABCDF, encoding serine proteases; purR, encoding pur operon repressor; set8 nm, encoding exotoxin NM8.