Abstract
The essential trace compounds tungstate and molybdate are taken up by cells via ABC transporters. Despite their similar ionic radii and chemical properties, the WtpA protein selectively binds tungstate in the presence of molybdate. Using site-directed mutagenesis of conserved binding pocket residues, we established a molecular basis for tungstate selectivity.
TEXT
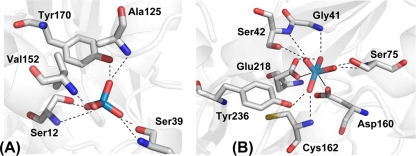
Molybdenum (Mo) and tungsten (W) have similar biochemistries (reviewed in reference 2). They are taken up by cells as the soluble oxoanions tungstate and molybdate via ATP-binding cassette (ABC) transporters. There are three types of tungstate/molybdate transporters, namely, ModABC (13), TupABC (11), and WtpABC (3), that differ both in protein sequence and in affinity for their respective periplasmic ligand binding proteins (Table 1). Isothermal titration calorimetry (ITC) displacement experiments with Pyrococcus furiosus WtpA showed its preference for tungstate in the presence of molybdate (3); however, the molecular basis of this observed selectivity is not understood. The two oxoanions have virtually identical ionic radii, due to the lanthanide contraction, and similar free energies of solvation (the calculated tungstate ΔGsolv is −226.9 kcal/mol, and the calculated molybdate ΔGsolv is −230.1 kcal/mol) (4). Crystal structures of WtpA in complex with tungstate or molybdate exhibited no significant differences (6, 7); however, they revealed an unusual octahedral W(VI)/Mo(VI)-binding motif that differs from the tetrahedral coordination observed in ModA structures (8, 10) and from an ill-defined half-open coordination in a TupA structure (5). In the structure of WtpA, two carboxylate oxygens from conserved glutamate (Glu218) and aspartate (Asp160) residues (P. furiosus WtpA numbering) directly interact with Mo/W, giving rise to an octahedral conformation (7) (Fig. 1). In this work, we used site-directed mutagenesis in combination with ligand affinity studies to gain insight into the molecular mechanism of the ligand selectivity of this class of periplasmic tungstate- and molybdate-binding proteins.
Table 1.
Dissociation constants of periplasmic binding proteins for molybdate and tungstatea
| Organism and protein (reference) |
KD for: |
KD displacement (nM) of: |
||
|---|---|---|---|---|
| Molybdate | Tungstate | MoO42− by WO42− | WO42− by MoO42− | |
| P. furiosus WtpA (3) | 11 ± 5 nM | <1 nM | 15 ± 4 | No displacement |
| E. acidaminophilum TupA (1) | ∼1.4 μM | ∼0.2 nM | ∼1 | No displacement |
| E. coli ModA (this work) | 0.13 ± 0.1 nM | 0.17 ± 0.1 nM | No displacement | No displacement |
Values are means and standard deviations.
Fig. 1.
Structures of periplasmic binding proteins in complex with tungstate indicating two binding modes. E. coli ModA (A) (Protein Data Bank file 1WOD [8]) binds tungstate in a tetrahedral conformation, whereas two additional bonds in P. furiosus WtpA (B) (Protein Data Bank file 3CG1 [6]) lead to an octahedral tungsten conformation. Broken lines indicate putative hydrogen bonds, and solid lines are covalent protein-metal bonds.
Ligand affinities of ModA.
ITC has been proven to be a sensitive method to determine affinity constants for tungstate- and molybdate-binding proteins, like WtpA and TupA, in the nanomolar and subnanomolar ranges (1, 3, 12). In order to relate the tungstate specificity of the WtpA protein to its unique octahedral binding coordination, we determined the specificity of Escherichia coli ModA, which binds oxoanions noncovalently in a tetrahedral conformation, with similar ITC experiments (Table 1). For this purpose, we cloned the modA gene into the pBAD/HisA vector (Invitrogen), expressed it in TOP10 cells, and purified it on Ni-Sepharose 6 fast-flow resin (GE Healthcare) via its N-terminal His tag. ITC experiments were performed as described in reference 3, with some adjustments. Tungstate and molybdate stock solutions of 200 μM were injected in 2- to 4-μl steps into the sample cell (1.42 ml) containing 4 to 10 μM ModA at 30°C in at least two independent experiments. The data obtained produced KD values that are over 2 orders of magnitude lower than the values reported for a previous determination using 99MoO42− in the absence or presence of unlabeled tungstate (9); however, they confirm that E. coli ModA is able to bind both tungstate and molybdate with very high affinities but is unable to replace one oxoanion with the other in a displacement titration (Table 1).
Ligand affinities of WtpA mutants.
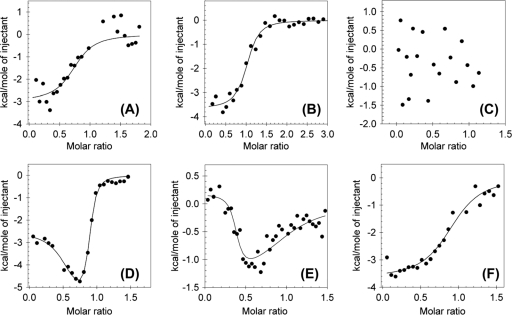
With the aim to study the role of the fully conserved Glu218 and Asp160 residues, both of which are involved in direct protein-to-metal bonds, we generated E218A, E218Q, D160A, and D160N mutant variants of WtpA using the QuikChange site-directed mutagenesis kit (Stratagene) with pBAD/HisA-WtpA as the template. WtpA mutant variants were expressed in TOP10 cells and purified as N-terminal His tag fusion proteins similar to ModA. ITC experiments performed as described in reference 3 showed that all four variants remain able to bind both molybdate and tungstate but with lower affinities than wild-type WtpA (Table 2). Looking at the aspartate mutant variants in more detail showed no large differences between the asparagine (D160N; ITC data not shown) and alanine (D160A) variants. Very prominent, however, was the lack of specificity of both of these variants; the D160A and D160N variants were no longer able to selectively bind tungstate, as born out by the lack of a signal in the displacement titration (Fig. 2A to C). This finding suggests that the presence of a direct bond between the metal and the carboxylate oxygen of aspartate 160 is essential for the protein's selectivity for tungstate.
Table 2.
Dissociation constants of wild-type and mutant WtpA at 40°Ca
| WtpA proteinb |
KD (nM) |
KD displacement (nM) | |
|---|---|---|---|
| MoO42− | WO42− | ||
| WT | 11 ± 5 | <1 | 15 ± 4 |
| D160N | 488 ± 126 | 415 ± 111 | No displacement |
| D160A | 193 ± 131 | 250 ± 215 | No displacement |
| E218Q | 1,613 ± 455 | 78 ± 20 | 329 ± 173 |
| E218A | 1,733 ± 832 (63%), 16 ± 12 (37%) | 64 ± 19 (40%), 7 ± 6 (60%) | 476 ± 126 |
| E218Ac | 46 ± 8 | 76 ± 15 | 1,449 ± 303 |
Values are means and standard deviations, which originated from the Origin fit, and percentages indicate distribution over two binding conformations.
Unless noted otherwise, pH 8.0.
pH 5.6.
Fig. 2.
Representative isothermal calorimetric titration curves of the D160A (A, B, C) and E218A (D, E, F) mutant variants of WtpA titrated with tungstate (A, D) or molybdate (B, E) and displacement titration of molybdate-saturated protein (2.5-fold excess) with tungstate (C, F) at 40°C. Filled circles represent the integrated heat calculated from the raw data (graphs not shown), and lines are fits obtained with the Origin software.
The E218A and E218Q glutamate variants, on the other hand, retained their tungstate specificity (Table 2; Fig. 2D to F), suggesting that this particular oxygen-metal interaction is not essential for the protein's selectivity. However, another interesting feature of the E218A variant can be noted; the shape of the binding curves indicates the presence of two binding sites with different affinities, which is not observed in the E218Q variant. This might be explained as follows. The WtpA crystal structure exposes a distance consistent with an additional interaction between E218 and tungstate/molybdate. Hollenstein et al. (6) proposed a single protonation of the oxoanion, which they describe, on the basis of observed bond lengths, as a shared proton bound via hydrogen bonds to the second oxygen of the E218 carboxyl group and to an oxygen atom of the oxoanion (Fig. 1B). In the E218Q variant, a similar hydrogen bond could be maintained through the amino group of the replacement glutamine side chain; however, in the alanine mutant, this interaction is not possible. We speculate that the glutamate-to-alanine mutation leads to alternative protein conformations with different affinities for the ligands. The E218A displacement titration data can be fitted into a one-binding-site model, suggesting that the protein conformations ultimately bind the oxoanion in the same manner. Furthermore, one of the binding sites is titrated away by reduction of the pH from 8.0 to 5.6, possibly via protonation of a residue involved in binding (Table 2).
In summary, these studies confirmed the hypothesis that the octahedral W(VI)/Mo(VI)-binding motif present in WtpA proteins is essential for its displayed binding preference for tungstate over molybdate. Mutation of aspartate 160 in a highly conserved DPxGYR motif (5) still allows the protein to bind both oxoanions with reduced affinity, but it completely abolishes the selectivity for tungstate. Mutation of fully conserved glutamate 218 only weakens the oxoanion binding. This study shows that the selectivity of the WtpA type of transporter depends on a single amino acid (D160) that is involved in coordination of the metal via a direct protein-metal bond.
Footnotes
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Andreesen J. R., Makdessi K. 2008. Tungsten, the surprisingly positively acting heavy metal element for prokaryotes. Ann. N. Y. Acad. Sci. 1125:215–229 [DOI] [PubMed] [Google Scholar]
- 2. Bevers L. E., Hagedoorn P.-L., Hagen W. R. 2009. The bioinorganic chemistry of tungsten. Coord. Chem. Rev. 253:269–290 [Google Scholar]
- 3. Bevers L. E., Hagedoorn P. L., Krijger G. C., Hagen W. R. 2006. Tungsten transport protein A (WtpA) in Pyrococcus furiosus: first member of a new class of tungstate and molybdate transporters. J. Bacteriol. 188:6498–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dudev T., Lim C. 2004. Oxyanion Selectivity in sulfate and molybdate transport proteins: an ab initio/CDM study. J. Am. Chem. Soc. 126:10296–10305 [DOI] [PubMed] [Google Scholar]
- 5. Hagen W. R. 2011. Cellular uptake of molybdenum and tungsten. Coord. Chem. Rev. 255:1117–1128 [Google Scholar]
- 6. Hollenstein K., et al. 2009. Distorted octahedral coordination of tungstate in a subfamily of specific binding proteins. J. Biol. Inorg. Chem. 14:663–672 [DOI] [PubMed] [Google Scholar]
- 7. Hollenstein K., Frei D. C., Locher K. P. 2007. Structure of an ABC transporter in complex with its binding protein. Nature 446:213–216 [DOI] [PubMed] [Google Scholar]
- 8. Hu Y., Rech S., Gunsalus R. P., Rees D. C. 1997. Crystal structure of the molybdate binding protein ModA. Nat. Struct. Biol. 4:703–707 [DOI] [PubMed] [Google Scholar]
- 9. Imperial J., Hadi M., Amy N. K. 1998. Molybdate binding by ModA, the periplasmic component of the Escherichia coli mod molybdate transport system. Biochim. Biophys. Acta 1370:337–346 [DOI] [PubMed] [Google Scholar]
- 10. Lawson D. M., Williams C. E., Mitchenall L. A., Pau R. N. 1998. Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 Å resolution crystal structure of Azotobacter vinelandii ModA. Structure 6:1529–1539 [DOI] [PubMed] [Google Scholar]
- 11. Makdessi K., Andreesen J. R., Pich A. 2001. Tungstate uptake by a highly specific ABC transporter in Eubacterium acidaminophilum. J. Biol. Chem. 276:24557–24564 [DOI] [PubMed] [Google Scholar]
- 12. Masters S. L., Howlett G. J., Pau R. N. 2005. The molybdate binding protein Mop from Haemophilus influenzae. Arch. Biochem. Biophys. 439:105–112 [DOI] [PubMed] [Google Scholar]
- 13. Rech S., Wolin C., Gunsalus R. P. 1996. Properties of the periplasmic ModA molybdate-binding protein of Escherichia coli. J. Biol. Chem. 271:2557–2562 [DOI] [PubMed] [Google Scholar]