Abstract
CodY is a global transcriptional regulator that is activated by branched-chain amino acids. A palindromic 15-bp sequence motif, AATTTTCNGAAAATT, is associated with CodY DNA binding. A gel mobility shift assay was used to examine the effect of pH on the binding of Bacillus subtilis CodY to the hutPp and ureAp3 promoters. CodY at pH 6.0 has higher affinity for DNA, more enhanced activation by isoleucine, and a lower propensity for nonspecific DNA binding than CodY at pH 8.0. DNase I footprinting was used to identify the CodY-protected regions in the hutPp and ureAp3 promoters. The CodY-protected sequences for both promoters were found to contain multiple copies of the 15-bp motif with 6-bp overlaps. Mutational analysis of the hutPp regulatory region revealed that two overlapping sequence motifs were required for CodY-mediated regulation. The presence of overlapping sequence motifs in the regulatory regions of many B. subtilis CodY-regulated genes suggests that CodY binds to native operators that contain overlapping binding sites.
INTRODUCTION
CodY is a global regulatory protein found in low-G+C Gram-positive bacteria that controls gene expression in response to nutrient availability (45). Microarray studies of different bacteria have shown that CodY regulates a large number of genes involved the adaptation to nutrient limitation (13, 20, 34). CodY has also been identified as a regulator of virulence in several pathogens (5, 15, 33). The Bacillus subtilis CodY protein has been shown to be a nutritional repressor of sporulation (39).
CodY is a dimeric protein that contains a winged helix-turn-helix DNA-binding motif and ligand-binding GAF domain (29). The activity of CodY is mediated by two different metabolic signals (19, 38, 39). Branched-chain amino acids bind to CodY proteins from many different bacterial species and stimulate their DNA-binding activities (14, 15, 20, 44). A subset of CodY proteins also respond to GTP. For instance, GTP binds to B. subtilis CodY and increases the DNA-binding affinity of this protein 4-fold (21, 39).
DNase I footprinting and mutational studies have established that a 15-bp DNA sequence motif, AATTTTCNGAAAATT, is associated with the binding of CodY to DNA (3, 13, 20). Base pair changes that decrease similarity with the consensus motif reduce CodY repression, while mutations that increase the similarity enhance the level of CodY-mediated regulation (3, 4, 14). Despite these experimental results, there are several unexplained observations about the DNA-binding specificity of CodY. For instance, the CodY-protected regions observed in DNase I footprinting experiments with the B. subtilis comK, dppA, flgBpA, hag, and srfA promoters lack sequences with high similarity to the consensus motif (3, 6, 42, 43). In addition, DNA sequences with high similarity to the CodY-binding motif are present in the upstream regulatory regions of many genes that are not subject to CodY-dependent regulation (3). Moreover, while a putative CodY-binding motif has been described for the hutP promoter (3, 13), a previously published study described a mutation outside this CodY-binding motif that relieved CodY-mediated regulation in vivo (16). Similarly, several mutations located outside the CodY-binding motif of the Lactococcus lactis oppD promoter have been shown to reduce the in vitro affinity of CodY (13, 14).
This communication describes the analysis of the B. subtilis CodY-regulated hutPp and ureAp3 promoters (18, 48). In this article, the term CodY DNA-binding site is used to refer to DNA sequences that correspond to the conserved 15-bp motif while the term CodY operator refers to the cis-acting DNA region where CodY regulates gene expression. The results from this study lead us to propose that CodY operators typically contain multiple CodY-binding sites with 6-bp overlaps.
MATERIALS AND METHODS
Gel mobility shift assays.
C-terminal His6-tagged CodY was overexpressed with plasmid pKT1 (7). CodY protein was purified by sequential chromatography on immobilized nickel affinity and size exclusion columns (7). The concentration of CodY was determined by measuring its UV absorbance at 280 nm. An extinction coefficient for His6-tagged CodY of 8,940 M−1 cm−1 was calculated from its amino acid sequence (37).
DNA fragments for the binding reactions were prepared by PCR amplification of plasmid DNA containing the hutPp and ureAp3 promoters. Plasmid pHUT724 contains a 200-bp EcoRV-NaeI hutPp promoter fragment (1). Plasmid pURE20 contains a 314-bp EcoRV-EcoRV ureAp3 promoter fragment (48). PCR products digested with EcoRI and HindIII were labeled by a fill-in reaction with [α-32P]dATP and Klenow DNA polymerase.
The DNA-binding reaction mixtures contained 0.1 nM end-labeled DNA fragment, 25 mM buffer, 200 mM sodium acetate, 1 mM dithiothreitol (DTT), 1 mM Tris(2-carboxyethyl)phosphine, 1 mM EDTA, 0.05% n-octylglucoside, 5% glycerol, 100 μg/ml bovine serum albumin, and 200 μg/ml sonicated calf thymus DNA. The binding reactions at pH 6.0, 7.0, and 8.0 used the buffers 2-(N-morpholino)ethanesulfonic acid, N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid, and Tris, respectively. The binding reaction mixtures were incubated at 30°C for 20 min and loaded onto 12% polyacrylamide gels with an acrylamide to bis-acrylamide ratio of 50 to 1. The gel electrophoresis buffers were designed to match the pH of the DNA-binding reaction mixtures, and each contained a pair of pK-matched buffer compounds with a concentration of 25 mM (30). The buffer pairs for electrophoresis at pH 6.0, pH 7.0, and 8.0 were 2-(N-morpholino)ethanesulfonic acid plus bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane, imidazole plus N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid, and triethanolamine plus N-[tris(hydroxymethyl)methyl]glycine, respectively. Following electrophoresis, the gels were dried and the radioactive bands determined by exposing the dried gel to a phosphorimage screen.
To quantitatively analyze the binding of CodY to DNA, the band intensities were determined with the volume measurement function of ImageQuant software (Molecular Dynamics). The binding curves had a sigmoidal shape, and thus nonlinear regression analysis was used to fit the data to the Hill equation θ = Ch/(Ch + K0.5h), where θ is the fraction of bound DNA, C is the total dimer concentration of CodY, K0.5 is the binding constant, and h is the Hill coefficient. While the DNA-binding activity of any single CodY preparation was reproducible, different preparations of CodY had different specific activities. The K0.5 values for the binding of CodY from various preparations to the hutPp promoter in the presence of 32 mM isoleucine ranged from 1 to 10 nM. Because of this behavior, all of the CodY DNA-binding constants were determined with a single CodY preparation.
DNase I footprinting.
The hutPp and ureAp3 DNA fragments for footprinting experiments were prepared by PCR amplification and digested with the restriction enzymes Acc65I and HindIII. The downstream ends of these promoter DNA fragments were labeled by a fill-in reaction with [α-32P]dATP and Klenow DNA polymerase. The footprinting reactions were performed at pH 6.0 in the presence of 32 mM isoleucine using the same conditions as the gel mobility shift assay except that the DNA fragment concentration was increased to 1 nM and the EDTA was replaced with 10 mM magnesium chloride and 2 mM calcium chloride. The binding reaction mixtures were incubated at 30°C for 20 min and then treated with DNase I for 2 min. The nuclease reactions were terminated by adding an equal volume of stop solution (40 mM EDTA, 40 mM EGTA, 1% SDS, 200 μg/ml tRNA). The samples were extracted with phenol-chloroform, precipitated with ethanol, and resuspended in formamide gel-loading buffer. Size standards of the end-labeled DNA fragments were prepared as previously described (31).
Oligonucleotide mutagenesis.
Site-directed mutagenesis of hutPp was performed by PCR overlap extension using pHUT724 as a template (22). The mutation-containing PCR DNA fragments were digested with EcoRI and HindIII and subsequently inserted into pJCD9 (10). The resulting plasmids were sequenced to confirm the presence of the desired mutation. Transcriptional lacZ fusions were constructed by inserting the mutant hutPp DNA fragments into pSFL1 (50). All lacZ fusions were integrated into the amyE gene of the isogenic B. subtilis strains 168 (trpC2) and SF168Y (trpC2 ΔcodY) (48).
Enzyme assays.
Methods for the cultivation of bacteria in the minimal medium of Neidhardt et al. (36) have been described previously (1). All cultures contained 0.5% glucose and 0.2% glutamine. The composition of the 16-amino-acid (16-aa) mixture present in some cultures has been described previously (1). β-Galactosidase was assayed in crude extracts prepared from cells grown to mid-log growth phase (1). The reported β-galactosidase levels were corrected for the endogenous activity present in cells containing the promoterless lacZ fusion vectors integrated into the amyE gene. One unit of β-galactosidase activity produced 1 nmol of o-nitrophenol per min.
RESULTS
Effect of pH on CodY DNA-binding activity.
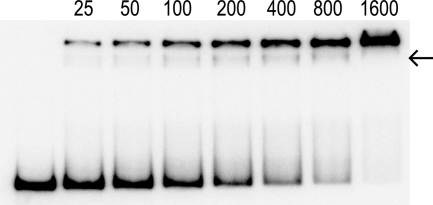
A gel mobility shift assay was used to examine the binding of CodY to a hutPp DNA fragment. The initial experiments were performed using conditions that were buffered at pH 8.0. Under these conditions, the CodY-hutPp interaction had a binding constant (K0.5) of 2,900 nM in the absence of any coeffectors and a K0.5 of 600 nM in the presence of 32 mM isoleucine (Table 1). These results demonstrated that CodY DNA-binding activity was enhanced 5-fold in the presence of isoleucine. Although this level of CodY DNA-binding enhancement is similar to that found in previously published reports (44, 46), it is rather low compared to that seen for other DNA-binding proteins. For example, the DNA-binding activities of the TrpR and PurR repressors are enhanced 200-fold by their corepressors (9, 51). In the pH 8.0 gel mobility shift autoradiographs, the CodY-DNA complexes form wavy bands, with tails at the ends of each band (Fig. 1). In addition, the mobility of the CodY-DNA complexes at pH 8.0 decreases as the protein concentration is increased. This decrease in complex mobility is presumably due to nonspecific binding of additional CodY dimers to the DNA fragment.
Table 1.
CodY DNA binding affinities
| Promoter | Coeffector(s)a |
K0.5 (nM) at pHb: |
||
|---|---|---|---|---|
| 6.0 | 7.0 | 8.0 | ||
| hutPp | None | 1,400 ± 100 | 2,600 ± 100 | 2,900 ± 100 |
| 32 mM Ile | 10 ± 1 | 400 ± 10 | 600 ± 10 | |
| 100 mM Ile | NDc | ND | 610 ± 10 | |
| 5 mM GTP | 1,500 ± 100 | ND | 2,500 ± 200 | |
| 32 mM Ile and 5 mM GTP | 13 ± 1 | 310 ± 10 | 420 ± 10 | |
| ureAp3 | None | 410 ± 20 | ND | 1,400 ± 100 |
| 32 mM Ile | 8.4 ± 0.3 | ND | 240 ± 10 | |
Isoleucine was present in the binding reaction mixture, gel, and running buffer. GTP was present only in the binding reaction mixture.
The experiments were performed with the binding reaction mixture, gel, and running buffer at the indicated pH. Values are the averages from two or more independent experiments. The uncertainty is the standard error from nonlinear regression analysis of the data.
ND, not determined.
Fig. 1.
CodY-hutPp gel mobility shift assays. These assays were performed at pH 8.0 (A) and at pH 6.0 (B). Isoleucine (32 mM) was included in the binding reaction mixture, gel, and gel buffer. The numbers above each lane correspond to the CodY dimer concentration (nM).
To minimize the non-sequence-specific DNA binding and obtain better isoleucine enhancement of the DNA-binding activity, the conditions for the CodY DNA-binding reactions were modified and tested. Replacement of the calf thymus nonspecific competitor DNA with synthetic copolymers, such as poly(dA-dT)·poly(dA-dT), poly(dG-dI)·poly(dG-dI), or poly(dA)·poly(dT), did not suppress the nonspecific DNA binding (data not shown). Similarly, reducing the sodium acetate concentration from 200 mM to 50 mM did not abate the CodY nonspecific DNA binding behavior (data not shown). Surprisingly, we found that lowering the pH to 6.0 had dramatic effects on CodY DNA-binding behavior. Experiments at pH 6.0 revealed that the K0.5 for CodY-hutPp binding was 1,400 nM in the absence of any coeffectors and 10 nM in the presence of 32 mM isoleucine (Table 1). Under these acidic conditions, isoleucine enhanced CodY DNA-binding activity 140-fold. In addition, at pH 6.0 the CodY-DNA complexes form tight bands and have uniform mobility, with CodY dimer concentrations that are 100-fold higher than the 10 nM binding constant (Fig. 1 and data not shown). These observations argue that the nonspecific DNA-binding activity of CodY is suppressed at pH 6.0. It is also noteworthy that in the presence of isoleucine CodY had a 60-fold-higher affinity for hutPp DNA at pH 6.0 (K0.5 of 10 nM) than at pH 8.0 (K0.5 of 600 nM). The hutPp DNA-binding behavior of CodY at pH 7.0 was very similar to its behavior at pH 8.0 (Table 1). GTP did not significantly enhance the hutPp DNA-binding affinity of CodY under the in vitro conditions used in these gel mobility shift assays (Table 1).
One possible explanation for the pH-dependent DNA-binding behavior of CodY is that the corepressor isoleucine might have a lower affinity for CodY at pH 8.0 than it does at pH 6.0. To test this hypothesis, the DNA-binding affinity of CodY for hutPp was examined at pH 8.0 in the presence of 100 mM isoleucine. We reasoned that if this hypothesis were correct, then increasing the concentration of isoleucine would increase the fraction of active CodY and thus result in an increase in the observed DNA-binding affinity. The experimental results showed that at pH 8.0 CodY had identical DNA-binding activities in the presence of 32 mM and 100 mM isoleucine (Table 1). These results are consistent with the idea that the lower DNA-binding affinity of CodY at pH 8.0 is not due to an inability of CodY to bind isoleucine at this alkaline pH.
The pH-dependent DNA-binding activity of CodY for a ureAp3 DNA fragment was also determined. The isoleucine-dependent enhancement of CodY affinity for ureAp3 DNA was 49-fold at pH 6.0 but only 6-fold at pH 8.0 (Table 1). In addition, the affinity of CodY for ureAp3 in the presence of 32 mM isoleucine was 29-fold higher at pH 6.0 (K0.5 of 8.4 nM) than at pH 8.0 (K0.5 of 240 nM). Taken together, these results argue that the enhanced in vitro DNA-binding affinity of CodY observed at pH 6.0 is a general effect that is not specific to a unique operator site.
Activation of CodY by different coeffectors.
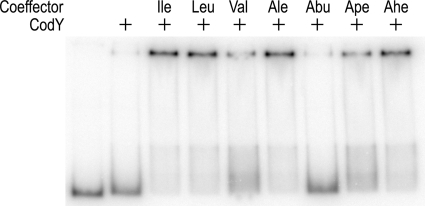
In the DNA-binding assays described above, the most stable CodY-DNA complexes were observed when 32 mM isoleucine was included in the binding reaction mixture, gel, and gel running buffer. Nonetheless, stable CodY-DNA complexes could also be generated when corepressor was added only to the binding reaction mixture (Fig. 2). This observation was exploited to examine the effectiveness of different aliphatic amino acids in activating the DNA-binding activity of CodY. These experiments used a fixed amount of CodY (150 nM) and varied the concentration of the corepressor. The fraction of bound DNA was quantified and plotted against the corepressor concentration. Nonlinear regression analysis was used to fit the data to the Hill equation and obtain a value for the effective concentration that gave rise to 50% DNA binding (EC50).
Fig. 2.
Enhancement of CodY DNA-binding activity by aliphatic amino acids. The experiments were performed at pH 6.0. CodY with a dimer concentration of 200 nM was present where indicated. All of the amino acids were l-stereoisomers and were present only in the binding reaction sample. Nonstandard abbreviations: Ale, alloisoleucine; Abu, 2-aminobutyric acid; Ape, 2-aminopentanoic acid; Ahe, 2-aminohexanoic acid.
Amino acids with 4-carbon side chains (leucine, isoleucine, alloisoleucine, and 2-aminohexanoic acid) were the most effective corepressors, with EC50s in the range of 4.7 to 6.8 mM (Table 2). The amino acids with 3-carbon side chains, 2-aminopentanoic acid and valine, required higher concentrations to activate CodY (EC50s of 17 and 22 mM, respectively). 2-Aminobutyric acid, which has only a 2-carbon side chain, was not a very effective corepressor (Table 2). These results are consistent with the idea that the size of the aliphatic side chain is a major determinant in the binding of amino acid corepressors to CodY.
Table 2.
Coeffector EC50 values for CodY DNA binding
| Coeffectora | EC50 (mM)b |
|---|---|
| Leucine | 4.7 ± 0.4 |
| Isoleucine | 4.9 ± 0.3 |
| Alloisoleucine | 5.5 ± 0.4 |
| 2-Aminohexanoic acid | 6.8 ± 0.3 |
| 2-Aminopentanoic acid | 17 ± 3 |
| Valine | 22 ± 3 |
| 2-Aminobutyric acid | >240c |
The l-stereoisomer of each amino acid was used for these experiments. The coeffectors were added only to the binding reaction mixture. These experiments were performed at pH 6.0 with 150 mM CodY dimer.
Each EC50 was determined from at least two independent experiments. The uncertainty is the standard error from nonlinear regression analysis of the data.
Only 30% of the DNA is bound with 240 mM 2-aminobutyric acid.
CodY-binding regions in hutPp and ureAp3.
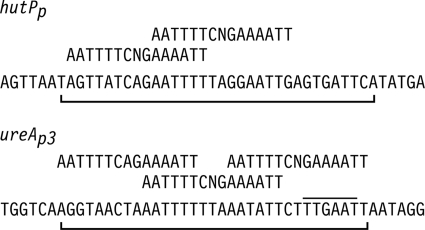
DNase I footprinting analysis was used to identify the CodY-protected regions in the hutPp and ureAp3 promoters. CodY protected sequences on the hutPp nontemplate DNA strand from positions +7 to +40 with respect to the transcriptional start site (Fig. 3). These results are consistent with an in vivo deletion analysis of the hutP promoter region which showed that DNA sequences between +20 and +42 are required for amino acid repression (50). The hutPp protected region contains two overlapping CodY DNA-binding site sequences with 4 mismatches to the consensus motif (Fig. 4).
Fig. 3.
DNase I footprinting of the hutPp and ureAp3 promoters with CodY. The molecular size markers are in the lanes labeled A+G. The CodY dimer concentration (nM) used for each reaction is indicated above each lane. All reactions were performed at pH 6.0 in the presence of 32 mM isoleucine. The protected regions are denoted by the vertical line.
Fig. 4.
Summary of the DNase I footprinting experiments. The nucleotide sequences of the nontemplate strands of the hutPp (+1 to +45) and ureAp3 (−66 to −22) promoters are shown. The −35 region of the ureAp3 promoter is overlined. The CodY-protected regions are indicated by the brackets below the DNA sequences. The CodY-binding consensus motifs are shown above the promoter sequences.
The nontemplate DNA strand of ureAp3 was protected from positions −60 to −28 (Fig. 3). The ureAp3 protected region contains three overlapping CodY DNA-binding site sequences with 5, 3, and 4 mismatches to the consensus motif (Fig. 4). The overlapping CodY DNA-binding sequences in each promoter have 6-bp overhangs.
Mutational analysis of the hutP promoter.
To analyze the contributions of the two CodY-binding sites in the hutP promoter to the regulation of gene expression from hutPp, mutations in the binding sites were generated by oligonucleotide-directed mutagenesis. Since the hut DNA promoter fragment used in the mutagenesis experiments lacks the downstream sequences required for catabolite repression and histidine induction, its expression is regulated only by CodY (18, 49, 50). The mutated promoters were transcriptionally fused to the lacZ gene and integrated into the B. subtilis chromosome as single copies. β-Galactosidase expression from the fusions was examined in cells grown in the presence and absence of the 16-aa mixture that promotes CodY-dependent repression (Table 3). In addition, the CodY DNA-binding affinities were determined with the gel mobility shift assay (Fig. 5).
Table 3.
Expression of hutPp-lacZ fusions in wild-type and ΔcodY strains
| hut promoter | β-Galactosidase sp act (U/mg of protein) in cells grown ona: |
Amino acid repression ratiob | CodY repression ratioc | ||
|---|---|---|---|---|---|
| Wild-type strain |
ΔcodY strain with Gln + 16 aa | ||||
| Gln | Gln + 16 aa | ||||
| Wild type | 6.6 | 0.4 | 18 | 17 | 45 |
| 11C | 12 | 8.1 | 14 | 1.5 | 1.7 |
| 12T | 1.4 | 0.01 | 21 | 140 | 2,100 |
| 14T | 13 | 7.0 | 22 | 1.9 | 3.1 |
| 16A | 19 | 17 | 18 | 1.1 | 1.1 |
| 18G | 20 | 16 | 19 | 1.3 | 1.2 |
| 24G | 12 | 1.1 | 17 | 11 | 15 |
| 25A | 14 | 2.0 | 17 | 7.0 | 8.5 |
| 26A | 0.08 | <0.01 | 17 | >80 | >1,700 |
| 28G | 13 | 7.4 | 17 | 1.8 | 2.3 |
| 30C | 5.2 | 0.23 | 18 | 23 | 78 |
Cells were cultured in glucose minimal medium containing the indicated nitrogen sources. All values are the averages of the results from two or more determinations. The standard errors were less than 20% for each value.
The amino acid repression ratio was calculated by dividing the β-galactosidase level for wild-type cells grown with glutamine by the value for wild-type cells grown with glutamine plus the 16-amino-acid mixture.
The CodY repression ratio was calculated by dividing the β-galactosidase level for ΔcodY cells grown with glutamine plus the 16-amino-acid mixture by the value for wild-type cells grown with glutamine plus the 16-amino-acid mixture.
Fig. 5.
Mutations in the hutPp promoter region. The hutPp DNA sequence from +8 to +31 is shown. The CodY-binding consensus motifs are displayed above the hutPp sequence. The CodY DNA-binding affinities were determined at pH 6.0 in the presence of 32 mM isoleucine and are shown in the rightmost column. The standard errors for the K0.5 values from nonlinear regression analysis of the binding data were less than 10% of each value. ND indicates binding affinities that were not determined.
Single base pair mutations that decreased the sequence similarity of the upstream CodY-binding site to the consensus motif, hutPp11C, hutPp14T, and hutPp16A, were found to reduce the level of repression by CodY in medium containing the 16-amino-acid mixture. DNA fragments containing these mutations had 4- to 7-fold-lower affinities for CodY than the wild-type DNA fragment.
Three mutations in the downstream CodY-binding site (hutPp25A, hutPp28G, and hutPp30C) that decreased the sequence similarity with the consensus motif were also constructed. The hutPp28G mutation relieved CodY-dependent regulation in vivo and reduced the in vitro CodY affinity 24-fold compared to that for wild-type hutPp. Expression from the hutPp25A promoter was only partially repressed by CodY in vivo, while the hutPp30C mutation had no significant effect on CodY-mediated repression.
The hutPp12T and hutPp26A mutations alter the upstream and downstream CodY-binding sites, respectively, so that each site has increased its similarity with the consensus motif (up mutations). Compared to the wild-type hutPp promoter, both of these up mutations increased the in vivo levels of CodY-dependent repression and increased the in vitro CodY DNA-binding affinities. Taken together, the results of these mutational studies demonstrate that both CodY-binding sites in the hutPp operator contribute to the in vivo regulation by CodY. The mutational analysis of hutPp by Eda et al. (16) also supports this conclusion.
CodY site up mutations can give rise to promoters that are essentially uninducible. For instance, the hutPp26A mutant promoter has a level of expression under nonrepressing conditions that is lower than the level observed with the hutPp wild-type promoter under repressing conditions (Table 3). The up mutations dppAp55, hutPp9A, and ylmAp1 have also been shown to be uninducible (3, 16). These results support the proposal that no naturally occurring CodY operators contain sequences identical to the consensus motif, because these promoters would be unable to derepress gene expression (3).
When analyzed with the gel mobility shift assay, only a single CodY-DNA complex was observed with the wild-type hutPp, wild-type ureAp3, and most of the hutPp mutant DNA fragments. The only exception was the hutPp28G DNA fragment, where a low-abundance CodY-DNA complex with intermediate mobility was observed (Fig. 6). It is noteworthy that the hutPp28G DNA fragment had the lowest in vitro CodY affinity of all of the hutPp mutant DNA fragments (Fig. 5). The presence of two CodY-DNA complexes in the hutPp28G gel shift experiments is consistent with the idea that at least two CodY dimers are present in the low-mobility CodY-hutPp28G complex.
Fig. 6.
Gel shift of the hutPp28G DNA fragment. This experiment was performed at pH 6.0 in the presence of 32 mM isoleucine. The intermediate band is indicated by the arrow. Numbers above the lanes are CodY dimer concentrations (nanomolar).
CodY-binding sites of B. subtilis genes.
A search of the DNA sequences upstream of B. subtilis CodY-regulated genes was performed to determine if additional CodY operators contain overlapping CodY DNA-binding sites. This search utilized a 24-bp tandem consensus sequence (TCS) that was composed of two CodY DNA-binding sites with a 6-bp overlap (Table 4). This search used a cutoff of 7 mismatches with the TCS.
Table 4.
CodY-binding sites in CodY-regulated B. subtilis genes
| Promoter region | No. of bp between site and downstream gene | DNA sequencea | No. of mismatches |
|---|---|---|---|
| CodY DNA-binding site 1 | ---------AATTTTCNGAAAATT | ||
| CodY DNA-binding site 2 | AATTTTCNGAAAATT--------- | ||
| Tandem consensus sequence | AATTTTCNGAAWWTTCNGAAAATT | ||
| Genes with a single TCS site | |||
| yurP | 86 | AtTaTTgAGAAATTTCAGAAAATa | 4 |
| glnQ | 37 | AATTTTCAGAAAAgTtTGAtcATT | 4 |
| ylmA | 29 | AgTTTTaTGAAAATTaAaAcAATT | 5 |
| bcaP | 167 | AATTTgtCGAtTTTTCTaAcAATT | 5 |
| ureAp3 | 867 | AATTTTtTaAATATTCTttgAATT | 5 |
| hutP | 2 | AgTTaTCAGAATTTTtAGgAAtTg | 6 |
| gabP | 25 | AATTaTCAtAATATTCAGtAAtga | 6 |
| flgB | 231 | AAagTTtCaAAAATgCCGAAAAga | 7 |
| rocR | 43 | AATagaaGcAATATTaAGAAAATa | 7 |
| appD | 98 | AATTTTtCGAtAATTCAatAttaT | 7 |
| ycgM | 89 | tATTTTgAGgATATTgTGAAcgcT | 7 |
| ilvA | 45 | tATTTTtTGAATATTCAtgttATa | 7 |
| acsA | 55 | tATaTTtTaAAAATTgAGAAgAaT | 7 |
| ycgA | 277 | cATaTTgCtAcTATTCAGAAtAaT | 7 |
| yhjC | 136 | tAaTTTCAGAcAATTCAcActATa | 7 |
| yurJ | 86 | tATTTTaGaAATATagCaAAAATg | 7 |
| Genes with two TCS sites | |||
| dppA | 11 | AATaTTCAtAATTTagTaAAAAag | 7 |
| dppA | 20 | AtTTgTtAGAATATTCAtAAttTa | 7 |
| ilvB | 521 | AATTgTCTaAtAATTtTaAAAAaT | 6 |
| ilvB | 530 | AtTaaTCAaAATTgTCTaAtAATT | 7 |
| yxbB | 414 | AATTaTCAGAggATTaTaAtAtTT | 7 |
| yxbB | 423 | tATTgaCAGAATTaTCAGAggATT | 6 |
| ureAp2 | 242 | tATTaTaGaAATTTcCAGAAAAaa | 7 |
| ureAp2 | 266 | AATTgTCGaAcTAgTCAGAcAAgT | 6 |
| ilvD | 41 | AATaaaCTGAAAATTgTcAAAATa | 6 |
| ilvD | 197 | AtTgTTgAcAAATaTCCGAAAAcT | 6 |
| ybgE | 104 | AtccTaaAGAATATTCTGAAAtTT | 6 |
| ybgE | 24 | AAaagaCTGAATATTtAaAcAATT | 7 |
| oppA | 44 | AtTTgTCTGAtATTTCTGAggATT | 5 |
| oppA | 93 | AATaaTCAGAAAAgaCGGAAcAga | 7 |
| yufN | 95 | AATTTTaTcgATTcTaAGAAAtTg | 7 |
| yufN | 106 | tATTaTCAGAAAATTtTatcgATT | 7 |
Bases with mismatches to the TCS are indicated in lowercase.
Among the genes with a single TCS site, the sequences identified for the bcaP, flgB, hutP, ureAp3, ylmA, and yurP promoters were found to overlap the CodY-protected regions from DNase I footprinting experiments (Fig. 4) (3, 4, 6). The TCS site upstream of gabP overlaps the promoters for this gene (17).
The rocABC and rocDEF operons were formerly shown to be regulated by CodY in microarray experiments (34). While no TCS sites were found upstream of the rocA or rocD gene, a single TCS site was found upstream of the rocR gene (Table 4). RocR is a transcriptional activator of the rocABC and rocDEF operons (8). These observations suggest that CodY may indirectly regulate expression of the roc operons by controlling the level of rocR expression.
Two TCS sites were found in the upstream DNA regions of several genes (Table 4). In three of these genes, dppA, ilvB, and yxbB, the TCS sites overlap to form a tandem array of three CodY-binding sites (Fig. 7) that was identical to the arrangement observed in the ureAp3 promoter (Fig. 4). The overlapping TCS sites in the dppA and ilvB promoters correspond to the CodY-protected sequences observed in DNase I footprinting experiments (43, 44). The presence of multiple CodY-binding sites in the dppA promoter region has been noted previously (3). The individual CodY-binding sites in the dppA promoter have relatively low sequence similarity with the CodY-binding consensus sequence (3). The ability of CodY to regulate dppA expression may be dependent on the tandem arrangement of the three CodY-binding sites shown in Fig. 7. The occurrence of nonoverlapping TCS sites for several genes may indicate the presence of two CodY operators. It has been shown that the regulation of bcaP by CodY is mediated by two independent operators (4).
Fig. 7.
Promoters with three tandem CodY-binding sites. The individual CodY-binding consensus motifs are shown at the top. Bases in the promoter region sequences with mismatches to the CodY-binding consensus motifs are indicated in lowercase.
CodY TCS sites are found within the experimentally identified ureAp2 and yxbB promoters (35, 48). Both of the TCS sites for the oppA gene are located downstream of the putative promoter for the opp operon (40). It is noteworthy that CodY is a nutritional repressor of sporulation and that opp gene mutants are defective in sporulation (39, 40). These observations suggest that the opp operon may be a target for the CodY-mediated repression of sporulation.
DISCUSSION
Several in vitro DNA-binding properties of CodY are significantly altered by the pH of the binding conditions. First, CodY has a higher affinity for DNA at pH 6.0 than at pH 8.0 (Table 1). Second, CodY DNA-binding affinity has a higher level of isoleucine enhancement at pH 6.0 than at pH 8.0 (Table 1). Third, there is a larger discrimination between sequence-specific and nonspecific DNA binding at pH 6.0 than at pH 8.0 (Fig. 1). The physiological significance of these observations is unclear because exponentially growing B. subtilis cells maintain an intracellular pH of ∼8.1 (32). The observation that Streptococcus mutans codY mutants are acid sensitive compared to wild-type cells (28) raised the possibility that enhanced CodY DNA binding at low pH might facilitate survival of B. subtilis cells under acidic growth conditions. However, no difference in growth of wild-type and codY B. subtilis cultures could be observed in low-pH growth medium (S. H. Fisher, unpublished data). Another explanation for this discrepancy is that the acidic in vitro conditions artificially activate the DNA-binding activity of CodY. This interpretation could indicate the existence of an additional in vivo corepressor for CodY.
The mutational analysis of hutPp clearly demonstrates that two overlapping CodY-binding sites are required for CodY-mediated regulation of this promoter. The presence of TCS sites within the CodY-protected regions of several other B. subtilis CodY-regulated promoters suggests that multiple CodY-binding sites are present in other B. subtilis CodY operators. Studies of DNA binding by CodY from other bacteria support the overlapping binding site model. For instance, a previously published bioinformatics analysis of Staphylococcus aureus CodY-binding DNA regions identified a 21-bp conserved motif (33). This motif corresponds to bases 3 to 23 of the 24-bp TCS. These observations suggest that many S. aureus CodY operators may also contain two CodY binding sites with a 6-bp overlap. In addition, mutational studies of the L. lactis oppD promoter are consistent with the idea that the CodY operator of this gene also contains two overlapping binding sites (14). It is tempting to speculate that this mode of DNA binding by CodY may be conserved in other bacteria.
Despite this evidence in favor of overlapping CodY-binding sites, the TCS has limited usefulness in identifying native CodY operators. For instance, in a search of the intergenic regions of the B. subtilis genome for TCS sites with seven or fewer mismatches, the majority of the identified sequences were upstream of genes that are not regulated by CodY (L. Wray, unpublished results). In addition, no TCS sites were identified in the CodY-protected region of the comK, flgBpA, hag, or srfA promoter (6, 42). Furthermore, the experimentally identified CodY operator located downstream of the B. subtilis bcaP promoter is not a good match to the TCS (4). A position-specific weight matrix based on the contributions of the individual base pairs to CodY-DNA binding would likely be more successful at identifying native CodY operators.
The binding of CodY to DNA appears to be highly cooperative. This assertion is supported by the finding that CodY forms a single complex in the gel shift experiments with wild-type hutPp DNA even though the mutational studies revealed that there are at least two CodY DNA-binding sites in the hutPp operator. Further support comes from the observation that the DNA-binding curves are sigmoidal. A cooperative binding mechanism, regardless of its molecular details, would be expected to have an optimal spacing of the CodY DNA-binding sites. The presence of sequences in many CodY operators that match the TCS suggests that a 6-bp overlap would be most favorable.
This DNA-binding model for CodY could explain the observed behavior in pH 8.0 gel shift experiments, where CodY-hutPp DNA complexes with progressively lower mobility are observed as the CodY protein concentration is increased (Fig. 1). This process could involve the iterative binding of CodY dimers to sequence-specific sites and subsequent binding to nonspecific sites. This hypothesis argues that the cooperative binding of CodY to DNA is less dependent upon sequence specificity at pH 8.0 than at pH 6.0. Similarly, the progressive binding of CodY dimers could explain the observed tendency of CodY to protect large extended regions of DNA in DNase I footprinting experiments (13, 42, 43, 44).
The proposed mechanism for CodY DNA binding has been observed with other repressors. The DtxR, Fur, QacR, TrpR, TtgR, and XylR proteins have been shown to interact with DNA at overlapping binding sites (2, 11, 24, 26, 27, 41, 47). The in vitro behavior of several of these proteins mimics that of CodY. For instance, the QacR repressor binds DNA cooperatively and forms a single QacR-DNA complex in gel mobility shift assays (41). In addition, the Fur repressor can protect large extended DNA regions in DNase I footprinting experiments (12, 25).
The TrpR repressor binds to its native operators with repressor dimer to DNA stoichiometries of 1:1, 2:1, and 3:1 (23). Similarly, Fur-DNA complexes with repressor dimer to DNA ratios of 1:1, 2:1, 3:1, and 4:1 have been reported (26). CodY may also bind to its native operators with different stoichiometries. As noted above, the dppA, ilvB, ureAp3, and yxbC promoters contain three overlapping CodY-binding sequences (Fig. 4 and 7). The CodY-protected region of the hutPp promoter is large enough to accommodate three CodY-binding sites (Fig. 4). The significance of this observation is unclear because the putative third CodY-binding site in hutPp would have 10 mismatches with the 15-bp consensus motif. Unfortunately, technical problems associated with the in vitro properties of CodY prevented the determination of CodY-DNA stoichiometries. Nonetheless, while the observations presented here strongly suggest that many CodY operators contain multiple overlapping CodY-binding sites, it is possible that some CodY operators may consist of only a single binding site. The ability of CodY dimers to bind to single or tandem sites would presumably be dependent on the DNA sequence context.
ACKNOWLEDGMENTS
This research was supported by Public Health Service grant GM051127 from the National Institutes of Health.
We thank A. L. Sonenshein for his kind gift of the CodY overexpression plasmid and both A. L. Sonenshein and B. Belitsky for helpful discussions.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Atkinson M. R., Wray L. V., Jr., Fisher S. H. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baichoo N., Helmann J. D. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belitsky B. R., Sonenshein A. L. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190:1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belitsky B. R., Sonenshein A. L. 2011. Contributions of multiple binding sites and effector-independent binding to CodY-mediated regulation in Bacillus subtilis. J. Bacteriol. 193:473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bennett H., et al. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453–1467 [DOI] [PubMed] [Google Scholar]
- 6. Bergara F., et al. 2003. CodY is a nutritional repressor of flagellar gene expression in Bacillus subtilis. J. Bacteriol. 185:3118–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blagova E. V., Levdikov V. M., Tachikawa K., Sonenshein A. L., Wilkinson A. J. 2003. Crystallization of the GTP-dependent transcriptional regulator CodY from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 59:155–157 [DOI] [PubMed] [Google Scholar]
- 8. Calogero S., et al. 1994. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J. Bacteriol. 176:1234–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carey J. 1988. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc. Natl. Acad. Sci. U. S. A. 85:975–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J. D., Morrison D. A. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64:155–164 [DOI] [PubMed] [Google Scholar]
- 11. Dahl M. K., Degenkolb J., Hillen W. 1994. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J. Mol. Biol. 243:413–424 [DOI] [PubMed] [Google Scholar]
- 12. de Lorenzo V., Wee S., Herrero M., Neilands J. B. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. den Hengst C. D., et al. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332–34342 [DOI] [PubMed] [Google Scholar]
- 14. den Hengst C. D., et al. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dineen S., Villapakkam A., Nordman J., Sonenshein A. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206–219 [DOI] [PubMed] [Google Scholar]
- 16. Eda S., Hoshino T., Oda M. 2000. Role of the DNA sequence downstream of the Bacillus subtilis hut promoter in regulation of the hut operon. Biosci. Biotechnol. Biochem. 64:484–491 [DOI] [PubMed] [Google Scholar]
- 17. Ferson A. E., Wray L. V., Jr., Fisher S. H. 1996. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol. Microbiol. 22:693–701 [DOI] [PubMed] [Google Scholar]
- 18. Fisher S. H., Rohrer K., Ferson A. E. 1996. Role of CodY in regulation of the Bacillus subtilis hut operon. J. Bacteriol. 178:377937–377984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guédon E., Serror P., Ehrlich S. D., Renault P., Delorme C. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227–1239 [DOI] [PubMed] [Google Scholar]
- 20. Guédon E., Sperandio B., Pons N., Ehrlich S. D., Renault P. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151:3895–3909 [DOI] [PubMed] [Google Scholar]
- 21. Handke L. D., Shivers R. P., Sonenshein A. L. 2008. Interaction of Bacillus subtilis CodY with GTP. J. Bacteriol. 190:798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the PCR. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 23. Jeeves M., Evans P. D., Parslow R. A., Jaseja M., Hyde E. I. 1999. Studies of the Escherichia coli Trp repressor binding to its five operators and to variant operator sequences. Eur. J. Biochem. 265:919–928 [DOI] [PubMed] [Google Scholar]
- 24. Krell T., et al. 2007. Optimization of the palindromic order of the TtgR operator enhances binding cooperativity. J. Mol. Biol. 369:1188–1199 [DOI] [PubMed] [Google Scholar]
- 25. Lavrrar J. L., Christoffersen C. A., McIntosh M. A. 2002. Fur-DNA interactions at the bidirectional fepDGC-entS promoter region in Escherichia coli. J. Mol. Biol. 322:983–995 [DOI] [PubMed] [Google Scholar]
- 26. Lavrrar J. L., McIntosh M. A. 2003. Architecture of a Fur binding site: a comparative analysis. J. Bacteriol. 185:2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lawson C. L., Carey J. 1993. Tandem binding in crystals of a trp repressor/operator half-site complex. Nature 366:178–182 [DOI] [PubMed] [Google Scholar]
- 28. Lemos J. A., Nascimento M. M., Lin V. K., Abranches J., Burne R. A. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190:5291–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levdikov V. M., Blagova E., Joseph P., Sonenshein A. L., Wilkinson A. J. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in Gram-positive bacteria. J. Biol. Chem. 281:11366–11373 [DOI] [PubMed] [Google Scholar]
- 30. Liu Q., Li X., Sommer S. S. 1999. pK-matched running buffers for gel electrophoresis. Anal. Biochem. 270:112–122 [DOI] [PubMed] [Google Scholar]
- 31. Liu S.-T., Hong G. F. 1998. Three-minute G + A specific reaction for DNA sequencing. Anal. Biochem. 255:158–159 [DOI] [PubMed] [Google Scholar]
- 32. Magill N. G., et al. 1996. Analysis of the relationship between the decrease in pH and accumulation of 3-phosphoglyceric acid in developing forespores of Bacillus species. J. Bacteriol. 178:2204–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Majerczyk C. D., et al. 2010. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192:2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Molle V., et al. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morinaga T., Kobayashi K., Ashida H., Fujita Y., Yoshida K. 2010. Transcriptional regulation of the Bacillus subtilis asnH operon and role of the 5′-proximal long sequence triplication in RNA stabilization. Microbiology 156:1632–1641 [DOI] [PubMed] [Google Scholar]
- 36. Neidhardt F. C., Bloch P. L., Smith D. F. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petranovic D., et al. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53:613–621 [DOI] [PubMed] [Google Scholar]
- 39. Ratnayake-Lecamwasam M., Serror P., Wong K. W., Sonenshein A. L. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rudner D. Z., LeDeaux J. R., Ireton K., Grossman A. D. 1991. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J. Bacteriol. 173:1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schumacher M. A., et al. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21:1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Serror P., Sonenshein A. L. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Serror P., Sonenshein A. L. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20:843–852 [DOI] [PubMed] [Google Scholar]
- 44. Shivers R. P., Sonenshein A. L. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599–611 [DOI] [PubMed] [Google Scholar]
- 45. Sonenshein A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207 [DOI] [PubMed] [Google Scholar]
- 46. Villapakkam A. C., et al. 2009. Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids. J. Bacteriol. 191:6865–6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. White A., Xiaochun D., vanderSpek J. C., Murphy J. R., Ringe D. 1998. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature 394:502–506 [DOI] [PubMed] [Google Scholar]
- 48. Wray L. V., Jr., Ferson A. E., Fisher S. H. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wray L. V., Jr., Fisher S. H. 1994. Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms: regulation of transcription initiation and inhibition of histidine transport. J. Bacteriol. 176:5466–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wray L. V., Jr., Pettengill F. K., Fisher S. H. 1994. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting located downstream of the transcription initiation site. J. Bacteriol. 176:1894–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu H., Moraitis M., Reedstrom R. J., Matthews K. S. 1998. Kinetic and thermodynamic studies of purine repressor binding to corepressor and operator DNA. J. Biol. Chem. 273:8958–8964 [DOI] [PubMed] [Google Scholar]