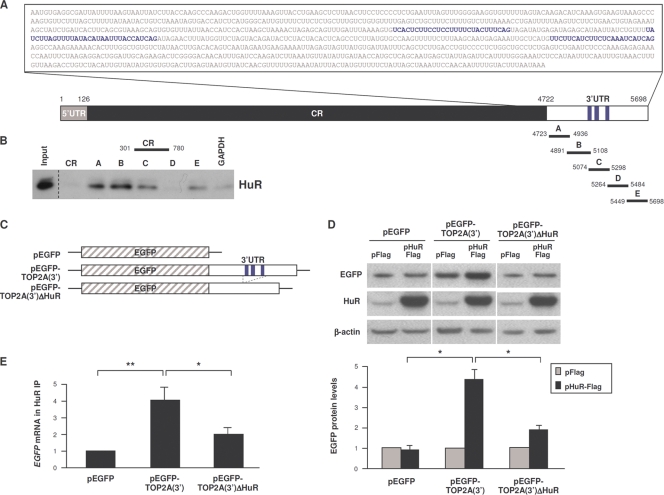
Fig. 2.
HuR binds to the TOP2A 3′UTR. (A) Schematic of the TOP2A 3′UTR depicting HuR interaction sites (blue text and stripes) identified by PAR-CLIP analysis (J. D. Keene, personal communication). Biotinylated transcripts (A to E) were synthesized spanning the TOP2A 3′UTR and a comparable segment of the TOP2A CR; a control biotinylated GAPDH 3′UTR RNA was used to measure background binding. (B) Incubation of equimolar biotinylated RNAs with whole-cell HeLa lysates was followed by pulldown of RNP complexes and HuR detection in the complexes by Western blot analysis. Input, 5 μg of whole-cell lysate. (C) Reporter constructs were engineered by cloning the TOP2A 3′UTR (full length or with deletion of the HuR CLIP sites) into plasmid pEGFP to generate pEGFP-TOP2A(3′) and pEGFP-TOP2A(3′)ΔHuR. (D) Cells were transfected with the plasmids shown in panel C together with either pFlag or pHuR-Flag; 16 h later, the levels of expressed EGFP protein were assessed by Western blot analysis (top), quantified by densitometry, normalized to α-tubulin levels, and plotted (bottom) (n = 5). (E) In cells that were transfected with the EGFP reporter plasmids shown in panel C, the association of HuR with the expressed transcripts [EGFP mRNA and chimeric EGFP-TOP2A(3′) and EGFP-TOP2A(3′)ΔHuR mRNAs] was measured 16 h later by RNP IP analysis. Shown are the means + SD (n = 3). *, P < 0.05; **, P < 0.01.