Abstract
Genomes of nucleocytoplasmic large DNA viruses (NCLDVs) encode enzymes that catalyze the formation of disulfide bonds between cysteine amino acid residues in proteins, a function essential for the proper assembly and propagation of NCLDV virions. Recently, a catalyst of disulfide formation was identified in baculoviruses, a group of large double-stranded DNA viruses considered phylogenetically distinct from NCLDVs. The NCLDV and baculovirus disulfide catalysts are flavin adenine dinucleotide (FAD)-binding sulfhydryl oxidases related to the cellular Erv enzyme family, but the baculovirus enzyme, the product of the Ac92 gene in Autographa californica multiple nucleopolyhedrovirus (AcMNPV), is highly divergent at the amino acid sequence level. The crystal structure of the Ac92 protein presented here shows a configuration of the active-site cysteine residues and bound cofactor similar to that observed in other Erv sulfhydryl oxidases. However, Ac92 has a complex quaternary structural arrangement not previously seen in cellular or viral enzymes of this family. This novel assembly comprises a dimer of pseudodimers with a striking 40-degree kink in the interface helix between subunits. The diversification of the Erv sulfhydryl oxidase enzymes in large double-stranded DNA viruses exemplifies the extreme degree to which these viruses can push the boundaries of protein family folds.
INTRODUCTION
In contrast to proteins that traverse the secretory pathway, cytosolic and nuclear proteins in mesophilic organisms rarely evolve to contain structural disulfide bonds. Some exceptions to this generalization are structural proteins encoded by nucleocytoplasmic large DNA viruses (NCLDVs), which do contain disulfides despite folding in an environment that typically is reducing (16). To promote disulfide formation in the cytosol or nucleus, NCLDVs (such as poxviruses, mimivirus, African swine fever virus [ASFV], iridoviruses, phycodnaviruses, and others) encode catalysts of disulfide formation (38, 40) similarly to cellular enzymes of the Erv (for essential for respiration and viability) family. The cellular Erv enzymes promote oxidative folding in the mitochondrial intermembrane space (30) and certain other biological settings (14, 41). Recently, an enzyme capable of catalyzing disulfide formation was identified in baculoviruses (28), a group of large double-stranded DNA viruses not classified as NCLDVs (21). The baculovirus sulfhydryl oxidase enzyme, one of a core group of conserved baculovirus gene products, is essential for virion assembly and thus for virus propagation (31, 48), similarly to its counterparts in NCLDVs (25, 39).
The sulfhydryl oxidase encoded by the Ac92 gene in the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) contains amino acid sequence features shared by other cellular and viral sulfhydryl oxidases (28). In particular, a CXXC motif at the amino terminus of a predicted helix corresponds to the active-site disulfide. A motif consisting of a tryptophan, three histidine, and two asparagine amino acid residues (WX3HXnHXmHNX2N) was predicted to contribute to the binding site for the flavin adenine dinucleotide (FAD) cofactor. Indeed, Ac92 was shown to be a flavoprotein (28).
A number of features, however, distinguish the baculovirus Ac92 enzyme from cellular and other viral sulfhydryl oxidases. First, Ac92 is predicted to comprise at least eight helices, whereas the canonical Erv enzyme fold contains only a four-helix bundle and a short fifth helix. The carboxy-terminal half of Ac92 corresponds to the predicted Erv family catalytic domain, but the structure and role of the amino-terminal region are unknown. In addition, large insertions were predicted between some of the helices in the Erv-like domain. Finally, Ac92 contains additional cysteines with unknown structural or functional roles.
In an earlier study of viral sulfhydryl oxidases, we observed diversity in the quaternary structural arrangements of the core catalytic module. In particular, the ASFV sulfhydryl oxidase has the tertiary structure of the Erv enzymes, but it assembles to form a dimer using an interface orthogonal to that of mimivirus and cellular homologs (17). Such variability in subunit packing contributes to differences in the shape and surface properties of the enzymes, as well as to differences in the relative orientations of the two active sites in the dimer. These differences may help define the substrates on which the enzymes act, the intracellular localization of the enzymes during virus assembly, and whether the enzymes ultimately are incorporated into virions or remain behind in the infected cell.
To determine how the divergent amino acid sequence of the baculovirus sulfhydryl oxidase affects the context and assembly of the core catalytic module, we determined the X-ray crystal structure of AcMNPV Ac92. Ac92 radically extends the structural diversity observed in the Erv enzyme family to date. We present here a structural description of Ac92 and a comparison with other viral and cellular sulfhydryl oxidases.
MATERIALS AND METHODS
Ac92 expression and purification.
The Ac92 open reading frame from AcMNPV was PCR amplified using the 5′ primer GGTCAGTACCATGGGCATACCGCTGACGCCGCTTTTTTCTC, the 3′ primer GTATGTCTCGAGGCCGCTGCTGCCGCGCGGCACCAGTTGCAAATTTAACAATTTTTTGTATTCTCCCC, and AcMNPV genomic DNA (a gift from Tamar Unger, Weizmann Institute of Science). PCR products were cloned into the pET-28a vector (Novagen) between the NcoI and XhoI restriction sites, such that a thrombin cleavage site was inserted upstream of the His6 tag at the carboxy terminus, which was encoded by the vector. Protein expression was done in the Escherichia coli strain BL21(DE3) (Novagen). Cells containing the expression plasmid were grown in LB containing 40 mg/liter kanamycin to an A595 of 0.5 to 0.6 at 37°C, isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mM, and the cultures were grown for a further 16 h at 25°C before harvesting. Cells were harvested and suspended in 50 mM sodium phosphate buffer, 500 mM NaCl, and 20 mM imidazole (pH 8.0) (buffer A) in the presence of protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride [PMSF] and 1 μg/ml each of leupeptin, aprotinin, and pepstatin A) and disrupted at 4°C by three passages through a French press at 11,000, which resulted in satisfactory cell lysis. The lysate then was ultracentrifuged for 30 min at 100,000 × g. The supernatant was collected and incubated with nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Qiagen) for 1 h at 4°C and loaded into a column. The resin was washed extensively with buffer A. Ac92 protein was eluted with 50 mM sodium phosphate buffer, 500 mM NaCl, and 250 mM imidazole (pH 8.0) (buffer B) and diluted to 50 mM sodium phosphate buffer, 250 mM NaCl, and 50 mM imidazole (pH 8.0). For His6 tag removal, the protein was incubated with thrombin for 8 h at room temperature. After PMSF was added to stop the cleavage reaction, the mixture was loaded onto Ni-NTA agarose resin preequilibrated with buffer A. Although the His6 tag was removed by the thrombin as determined by Western blotting using anti-His5 antibodies (Qiagen), the cleaved protein nevertheless bound the Ni-NTA resin in the presence of 50 mM imidazole. The protein was eluted with buffer B, concentrated, and further purified by size-exclusion chromatography (Superdex 75; GE Healthcare) in 50 mM sodium phosphate buffer, 250 mM NaCl, and 50 mM imidazole (pH 7.8). Ac92 was concentrated to 12 to 14 mg/ml as determined spectroscopically at 446 nm (ε = 11,300 M−1 cm−1, which corresponds to absorbance from the FAD) in 6 M guanidine HCl, 20 mM sodium phosphate buffer (pH 6.5). A selenomethionine variant was prepared according to a published protocol (44) and purified as described for wild-type Ac92.
Crystallization, data collection, and structure refinement.
Using the hanging drop vapor diffusion method, Ac92 crystallized readily in many polyethylene glycol (PEG)-ion solutions. These crystals had a plate-like morphology, belonged to the P1 space group, and suffered from high mosaicity. The best crystals of this form were grown over a well solution containing 0.1 M bicine (pH 8.5), 50 mM arginine HCl, 5 to 7% dimethylsulfoxide (DMSO), 1 mM ZnCl2, and 4 to 10% (wt/vol) PEG 2000 monomethyl ether. An extensive search for a different space group yielded large block-like crystals in space group C2, which produced higher quality diffraction data (Tables 1 and 2). C2 crystals and a selenomethionine variant were grown over a well solution containing 0.1 M acetic acid (pH 4.4), 50 mM arginine HCl (pH 5.7), 750 mM NaCl, 5% ethanol, and 22 to 24% (wt/vol) PEG 5000 monomethyl ether. Crystals of both the P1 and C2 forms, in their respective mother liquors, were mixed with an equal volume of 20 mM sodium phosphate buffer (pH 6.5), 20% PEG 1500, 20% ethylene glycol, 15% isopropanol, and 10% dimethyl sulfoxide before flash-freezing in liquid nitrogen.
Table 1.
Summary of crystallographic data statistics
| Parameter | Value for Ac92 crystalsc |
||||
|---|---|---|---|---|---|
| Native | MAD peak | MAD inflection | MAD remote | Native | |
| Space group | C2 | C2 | C2 | C2 | P1 |
| Wavelength (Å) | 0.97692 | 0.97922 | 0.97937 | 0.97623 | 0.97692 |
| Unit cell dimensions (Å) | |||||
| a | 55.76 | 55.68 | 55.59 | 55.55 | |
| a = b | 50.73 | ||||
| b | 80.83 | 80.46 | 80.86 | 80.43 | |
| c | 65.05 | 65.04 | 64.91 | 65.01 | 65.17 |
| α | 83.95 | ||||
| α = γ | 90.00 | 90.00 | 90.00 | 90.00 | |
| β | 105.955 | 105.90 | 105.79 | 105.88 | 79.01 |
| γ | 65.58 | ||||
| Resolution (Å) | 50–1.47 (1.5–1.47) | 33.84–2.14 (2.26–2.14) | 62.46–2.14 (2.26–2.14) | 62.53–2.14 (2.25–2.14) | 40.34–2.14 (2.26–2.14) |
| Completeness (%) | 99.1 (99.1) | 99.7 (99.6) | 99.8 (99.6) | 99.9 (99.7) | 97.6 (95.7) |
| Redundancy | 6.8 (5.7) | 7.2 (7.0) | 7.2 (7.0) | 7.2 (7.0) | 3.8 (3.8) |
| Rsyma | 0.069 (0.958) | 0.056 (0.125) | 0.053 (0.127) | 0.058 (0.160) | 0.075 (0.399) |
| I/σ | 19.2 (2.5) | 22.7 (13.7) | 22.4 (13.7) | 23.2 (13.4) | 8.5 (3.1) |
| Overall figure of meritb | 0.661/0.817 | ||||
Rsym = ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl), where Ii(hkl) is the observed intensity and <I(hkl)> is the average intensity for i observations.
Figures of merit for data from 20- to 2.14-Å resolution were obtained from Phenix (2).
Numbers in parentheses refer to the highest-resolution shell.
Table 2.
Summary of refinement statisticsa
| Parameter | Refinement statistics for space group: |
|
|---|---|---|
| C2 | P1 | |
| Total reflections/test set | 46,366/3,259 | 31,277/2,183 |
| Rwork/Rfree | 20.8/23.3 | 23.0/26.9 |
| RMS deviations from ideality | ||
| Bond (Å) | 0.00538 | 0.00739 |
| Angle (°) | 0.95014 | 1.04432 |
| No. of atoms | ||
| Protein | 2,324 | 4,098 |
| Water | 214 | 139 |
| FAD | 73 | 146 |
| Imidazole | 5 | 5 |
| Acetate | 4 | |
| Phosphate ion | 10 | |
Rwork/Rfree = Σ||Fobs| − |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. A set of reflections (7.1%) were excluded from refinement and used to calculate Rfree. RMS, root mean square.
Diffraction data were collected on the ID23-1 beamline at the European Synchrotron Radiation Facility (ESRF), Grenoble, France, using an ADSC Q315R charge-coupled device (CCD) detector. Native data were integrated and scaled using HKL2000 (33). For phase determination, multiwavelength anomalous dispersion data were collected using a selenomethionine crystal of space group C2. These data sets were processed and scaled using the MOSFLM (4) and SCALA (1) programs. Heavy atom positions were located, and phases were calculated using Phenix (2). Model building and refinement were done using COOT (12) and CNS (6), respectively. Depictions of protein structures were made using PyMOL (www.pymol.org). The structure in the P1 crystal form was determined by molecular replacement using Phaser (29), with a dimer of the C2 structure generated by applying crystallographic symmetry as the search model. Maps calculated from the molecular replacement solution showed discrepancies within one of the subunits of the dimer, so maps were recalculated using phases derived from a model lacking four helices of that subunit. The helices then were rebuilt into the new maps prior to cycles of refinement and more modest rebuilding.
Analytical ultracentrifugation.
The analytical ultracentrifugation of Ac92 was performed in 50 mM sodium phosphate buffer, 250 mM NaCl, and 50 mM imidazole (pH 7.8) at protein concentrations in the range of 5 to 20 μM. Absorbance data (280 and 446 nm) were collected at 20°C at 4-h intervals at speeds of 9,000, 10,000, 11,000, 14,500, 15,500, 16,500, 17,500, and 18,500 rpm in a Beckman Optima XL-A centrifuge equipped with an AN50 Ti rotor and a six-channel centerpiece. The equilibrium data were analyzed with the nonlinear least-squares fitting program UltraScan (11) using an ideal, single-component model for the global fit.
Protein structure accession numbers.
The atomic coordinates and structure factors were deposited in the Protein Data Bank (PDB) (www.rcsb.org). The PDB codes are 3P0K and 3QZY for the C2 and P1 crystal forms, respectively.
RESULTS
Ac92 structure determination.
AcMNPV Ac92 was crystallized in two space groups, C2 and P1. Diffraction data were collected to 1.47 and 2.14 Å, respectively. Phases were determined by multiwavelength anomalous dispersion using a C2 crystal of Ac92 replaced with selenomethionine. A preliminary model of the Ac92 dimer, generated from one protomer using crystallographic symmetry, was used to solve the P1 crystals by molecular replacement.
A novel dimer configuration in Ac92.
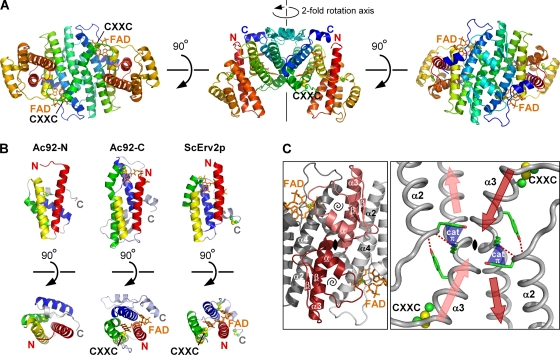
Ac92 forms a large, helical dimer of ∼63 kDa (Fig. 1 A). Each subunit in the dimer consists of two four-helix bundles. The amino-terminal bundle (Ac92-N) has the same topology as the Erv fold but exhibits different interhelical packing angles (Fig. 1B) and does not bind a cofactor. In contrast, the carboxy-terminal bundle (Ac92-C) is a true Erv family sulfhydryl oxidase domain. It binds FAD, which is consistent with the presence of the WX3HXnHXmHNX2N motif, and contains an FAD-proximal disulfide as found in other functional sulfhydryl oxidases (Fig. 1B).
Fig. 1.
Ac92 exhibits a new quaternary structure for Erv enzymes. The FAD is shown as orange sticks, and cysteine side chains are balls and sticks. The CXXC label indicates the position of the active-site dicysteine motif adjacent to the FAD. (A) Each polypeptide chain is colored from red (amino terminus; N) to blue (carboxy terminus; C). Ac92 dimerizes via the carboxy terminus of its two helical bundle domains. The three views of the dimer are related by successive rotations around the horizontal axis. (B) Both domains of Ac92 have the helical bundle topology of the Erv family enzymes. The two helical bundles of the Ac92 protomer (Ac92-N and Ac92-C are the amino- and carboxy-terminal bundles, respectively) are compared to the structure of a subunit of a cellular Erv family sulfhydryl oxidase, the S. cerevisiae Erv2 protein (ScErv2p; PDB code 1JR8). The four primary helices in each domain are colored red, yellow, green, and blue in order from amino to carboxy terminus. Two views of each domain, related by rotation around the horizontal axis, are shown. (C) The Ac92 dimer interface has a unique geometry. The 2-fold axis of the dimer runs through the black symbol in the center of each panel. In the left panel, one subunit is light gray and the second is dark gray, with the baculovirus-specific insertions in pink and red. Within these insertions, the secondary structural elements of the β-α-β motif are labeled (underlined symbols), and a spiral symbol is placed within the loop enclosed by this motif. The view in the right panel was obtained by rotating 180° around the vertical axis and zooming in. The red and pink arrows indicate the amino- to carboxy-terminal directionality of the helices, and the longer arrows emphasize the collision course of the α3 helices prior to the kink. Putative hydrogen bonds are shown as red dashed lines, and a potential cation-π interaction stabilizing the geometry in this region is represented with a blue cone labeled cat π.
Ac92 dimerizes via its FAD binding domain (Ac92-C), as do other Erv sulfhydryl oxidases with known structures (10, 15, 17, 45, 47). However, in contrast to cellular Erv enzymes and predictions for most viral sulfhydryl oxidases, Ac92 dimerization occurs using a different surface of the FAD-binding domain. Such a phenomenon was previously observed in the structure of the ASFV sulfhydryl oxidase pB119L (17), but the Ac92 quaternary structural arrangement bears no similarity to that observed for the ASFV enzyme. In particular, the standard Erv interface buries helices α1 and α2 of the four-helix bundle, the Ac92 dimer interface involves helices α3 and α4 of Ac92-C, and ASFV pB119L uses α2 and α3 for self association. As discussed further below, Ac92 thus represents a third mode for the self assembly of Erv sulfhydryl oxidases.
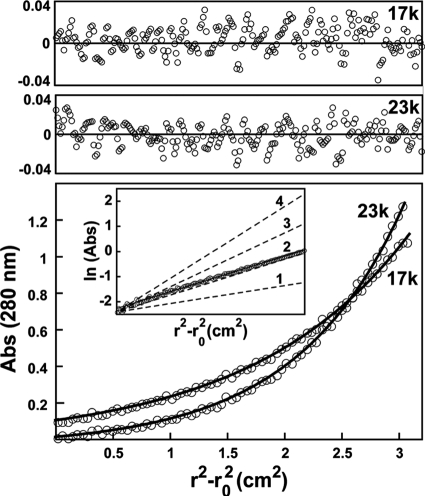
The Ac92 dimer interface, in addition to involving a different surface, also is expanded due to the elaboration of certain secondary structural elements and the incorporation of new motifs. In particular, an insertion consisting of a dramatic extension of helix α3 followed by an additional β-α-β motif contributes to dimerization (Fig. 1C, left). The long α3 helix is the most striking feature of the dimer interface. The first 10 residues downstream of the active-site CXXC motif form a helix that extends on a collision course with its symmetry mate, but the helix then kinks (Fig. 1C, right). The portion of helix α3 downstream of the kink interacts with the carboxy-terminal half of helix α4 of the second subunit in the dimer. This same interface is seen in both the P1 and C2 crystal structures and is largely hydrophobic. Ac92 is a dimer in solution (Fig. 2), as determined by analytical ultracentrifugation, further supporting the likelihood that the self association observed crystallographically occurs through an evolved interface. The Ac92 dimer buries approximately 1,100 Å2 of surface area, while a range of 590 to 760 Å2 is seen for other cellular and viral Erv enzyme dimers (17).
Fig. 2.
Analytical equilibrium ultracentrifugation shows Ac92 to be a dimer in solution. The measured solution molecular mass was 59 kDa; it was calculated to be 63 kDa based on amino acid sequence. In the bottom panel, two representative 280-nm absorbance traces, taken at 17,000 and 23,000 rpm from a sample at approximately 10 μM protomer concentration, are shown as open circles. The results of the global fit to the entire data set (see Materials and Methods) are shown as black curves. The upper two panels show the residuals between the data and the fit for each of these two traces. The inset in the lower panel shows that the slope of the plot of the natural logarithm of the absorbance versus the square of the radius corresponds to that calculated for a dimer. Calculated slopes for monomeric (1), dimeric (2), trimeric (3), and tetrameric (4) species are indicated.
Ac92 contains a pseudodimer of Erv-like domains.
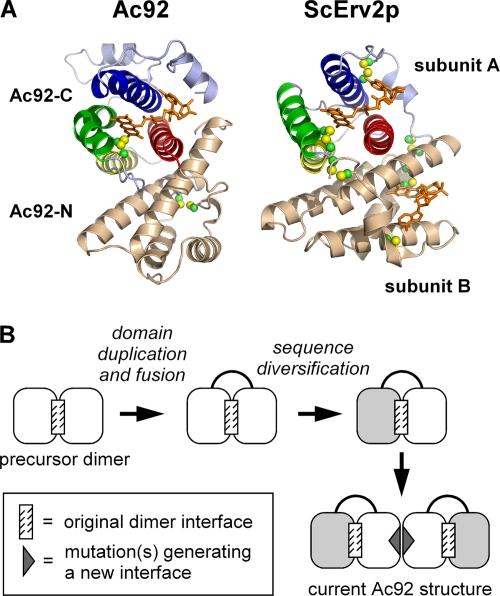
The overall quaternary structural arrangement of Ac92 described above is unprecedented for an Erv sulfhydryl oxidase, but the relative orientation of the two four-helix bundles within a single Ac92 protomer is strikingly similar to the arrangement of subunits in cellular Erv enzyme dimers (Fig. 3). Helices α1 and α2 of each Ac92 bundle form the intramolecular, interdomain interface, and the packing angle of α1/α2 with their pseudosymmetry mates is approximately 55°, which is the same packing geometry previously observed between the two subunits of single-domain Erv homodimers (10, 15, 17, 45, 47). It appears that the Ac92 precursor underwent gene duplication and fusion, preserving the original dimer interface in the interaction of the fused domains. The dimerization of the resulting two-domain pseudodimer then presumably generated the large assembly observed in the Ac92 structure (Fig. 1A). This model, in which the apparent gene duplication and fusion in Ac92 preceded the evolution of the interface involving helices α3 and α4, is most likely, since the mutation of the precursor homodimer to form the additional α3/α4 interface would have created an open symmetry, i.e., the possibility of polymerizing indefinitely. In contrast, if gene duplication and fusion occurred first and the two fused domains then diverged in sequence from one another, a mutation within one domain that promoted a new dimer interface would not affect the behavior of the second domain and would result in a closed symmetry with a defined number of subunits (Fig. 3B).
Fig. 3.
Pseudosymmetry in Ac92 suggests an evolutionary pathway for the formation of its novel quaternary structure. (A) One subunit of the Ac92 dimer is compared to a homodimer of S. cerevisiae Erv2p (ScErv2p). The redox-active domain of Ac92, in various colors, packs against the redox-inactive domain, in beige, in a manner similar to how the two subunits of dimeric cellular Erv enzymes self associate. In the ScErv2p structure, one subunit is shown in various colors and the second in beige. (B) Proposed evolutionary route to the Ac92 complex architecture. A homodimeric precursor may have undergone domain duplication and fusion to generate a single-chain pseudodimer. One of the domains then lost activity (light gray). The mutation of the redox-active domain (dark gray triangle) may then have enabled the dimerization of the pseudodimer.
The pseudosymmetry within each Ac92 subunit is reminiscent of a similar phenomenon found in enzymes of the quiescin sulfhydryl oxidase (QSOX) family. QSOX enzymes contain at the amino terminus either one or two thioredoxin fold domains, followed by two domains with the fold of the Erv enzyme family. Just as in Ac92, the first QSOX Erv-like domain lacks catalytic residues and a cofactor, whereas the second is a functional sulfhydryl oxidase module that packs against the preceding domain in the same manner as the two subunits of single-domain Erv enzyme dimers. Unlike baculovirus Ac92, however, QSOX enzymes are monomeric (18); the pseudodimer of degenerate and active Erv folds in QSOX does not further self-assemble.
The Ac92 active site.
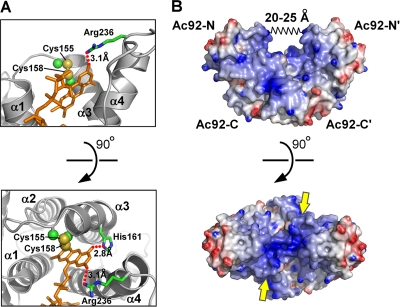
The Ac92 active site (Fig. 4 A) has many features in common with other Erv enzymes, e.g., the FAD assumes a similar configuration, and the active-site disulfide (Cys 155-Cys 158) is positioned abutting the FAD isoalloxazine as is typical in sulfhydryl oxidases. However, there are also a number of notable differences. First, helix α1 in Ac92-C is shorter than usual (red helix in Fig. 1B). In other Erv enzymes, e.g., yeast Erv2p (15) and Arabidopsis thaliana Erv1 (45), helix α1 extends above the active site and projects a basic side chain over the active-site disulfide in the direction of the FAD O2 atom. This basic residue may help stabilize negatively charged forms of the FAD and facilitate disulfide bond formation at the neighboring cysteine residues. In Ac92, an analogous interaction with the FAD O2 atom is provided by a basic residue arising not from helix α1 but rather from the carboxy terminus of helix α4 (Arg 236) (Fig. 4A). From this position, Arg236 approaches the FAD without draping over the active-site cysteines. The effect of these differences is to increase the apparent steric accessibility of the active-site disulfide of Ac92 compared to those of other Erv family sulfhydryl oxidases. Interestingly, in the ASFV sulfhydryl oxidase, a basic residue capable of extending into the vicinity of the FAD O2 atom arises from helix α3, between the two active-site cysteines in the CXXC motif. The viral sulfhydryl oxidases thus emphasize that important chemical and physical features contributing to enzyme function may be conserved spatially but not at the amino acid sequence level.
Fig. 4.
Ac92 active-site and surface properties. (A) The active-site region of Ac92 is shown with the side chain atoms of the CXXC motif in spheres at half their van der Waals radii. The FAD is in orange sticks, and the side chains of residues making key electrostatic interactions (dotted red lines) with the FAD also are shown as sticks. (B) An electrostatic surface representation of the Ac92 dimer is shown with regions of basic potential colored blue (>10 kBT/e) and acidic regions red (<−10 kBT/e, where kB is the Boltzmann constant, T is the absolute temperature, and e is the electron charge). The spring labeled 20 to 25 Å in the top panel represents the variability seen in the distance between the walls of the groove in the Ac92 structure when the P1 and C2 crystal structures were compared. The approximate locations of the Ac92-N and Ac92-C domains and their symmetry-related copies (Ac92-N′ and Ac92-C′) are indicated. In the bottom view, yellow arrows point at the active-site disulfides and approximate the direction from which substrate thiols would approach.
Surface properties of Ac92.
The two active sites of the Ac92 dimer are positioned at the two opposite entrances to a valley within the protein surface topography. The walls of the valley are formed by Ac92-N and the base by Ac92-C (Fig. 4B, top). More specifically, the valley is lined on either side by the first two helices of Ac92-N and on the bottom by helix α2 and the amino-terminal end of the long, kinked α3 helix of Ac92-C. The valley walls are hydrophobic or weakly basic, and the valley floor is more intensely positively charged. The location and geometry of the active-site disulfides imply that a substrate cysteine would have to approach from outside as opposed to from within the valley (Fig. 4B, bottom). The two snapshots of Ac92 obtained from the two crystal forms imply that the flexion of the Ac92-N domains leads to changes on the order of 5 Å in the width of the top of the valley (Fig. 4B, top).
Additional cysteine residues in Ac92.
The amino-terminal pseudo-Erv bundle, Ac92-N, contains two cysteine residues, C50 and C97. These cysteines are close in space to one another and point into the core of the bundle, but they are not linked in a disulfide in our structure. It remains to be determined whether cysteines C50 and C97 form a disulfide in infected cells, are redox active, or have another functional role in Ac92. Interestingly, these cysteines are at approximately the same position within the Ac92-N helical bundle as the FAD isoalloxazine in Ac92-C. It should be noted, however, that another hydrophobic residue replaces the cysteine corresponding to C97 in many baculoviruses, so phylogenetics does not suggest a structurally or functionally essential role for this cysteine pair.
DISCUSSION
The Erv family sulfhydryl oxidases are found throughout eukaryotic species and play an important role in protein localization to mitochondria and the assembly of the respiratory chain (3, 27). In addition, cytosolic and secreted forms have been shown to have particular activity as liver-specific growth and regeneration factors in mammals (34). These cellularly encoded Erv enzymes typically are small, single-domain dimers with at most a flexible amino- or carboxy-terminal extension involved in organelle targeting, membrane localization, or substrate interactions (9, 13, 43, 45). The only known examples of cellular proteins with an Erv fold functioning in a multidomain context are enzymes of the QSOX family, in which an Erv domain is fused downstream of a domain or domains having the thioredoxin (trx)-fold (7). In particular, the first trx domain of some QSOX orthologs has recognizable homology to the redox-active trx-fold domains of protein disulfide isomerase (PDI), an oxidoreductase involved in disulfide formation and rearrangement in the endoplasmic reticulum. Within QSOX, the Erv and PDI-like domains cooperate in the generation and transfer of disulfides to substrate proteins (24).
Although Erv sulfhydryl oxidases encoded by viruses generally are small, compact proteins like most cellular Erv enzymes, exceptions are found, for example, in ascoviruses, mimivirus, and baculoviruses. The Erv sulfhydryl oxidases of ascoviruses and mimiviruses, both NCLDVs, have carboxy-terminal structured extensions. The baculovirus Erv sulfhydryl oxidase, Ac92, has an additional domain fused amino terminally to the FAD-binding, sulfhydryl oxidase domain. Unlike in QSOX, however, the additional domains in these virus disulfide catalysts are not homologous to other known proteins, so it was not clear whether these domains have structural, functional, or other roles.
The X-ray crystal structure of baculovirus Ac92 suggests that the amino-terminal domain of this protein does not serve a role comparable to that of the trx domain but rather to the degenerate Erv domain of QSOX. Although Ac92-N has two cysteines in close proximity in the tertiary structure, these cysteines are not appropriately positioned to undergo dithiol/disulfide rearrangement with other proteins or with the FAD-proximal disulfide in the Ac92-C domain within either the same or the adjacent Ac92 subunit. Furthermore, the finding that these cysteines are unpaired in the crystals indicates that they were not oxidized by electron transfer to the FAD-proximal disulfide. In QSOX, the redox-active trx-fold region is flexibly tethered to the Erv domain (A. Alon and D. Fass, unpublished data) (35), presumably to enable interaction first with the substrate and then with the FAD-containing active site. The modest range of motion in Ac92 suggested by the comparison of the P1 and C2 crystal structures, as well as the presence of a well-packed hydrophobic interface between Ac92-N and Ac92-C, does not support the likelihood of major domain rearrangements as part of the Ac92 catalytic cycle. The Ac92-N helical bundle thus most likely has a stabilizing, structural role. It also may contribute to enzyme localization or substrate recognition by providing additional binding sites (31).
Baculovirus Ac92 is composed of both an intramolecular pseudodimer (Fig. 3) as well as an intermolecular true dimer (Fig. 1A), making it conceptually different from the dimer interface migration observed previously for ASFV pB119L. In ASFV pB119L, the canonical interface between helices α1/α2 and their symmetry-related helices is absent and is replaced by an alternative interface involving helices α2/α3. In baculovirus Ac92, the original α1/α2 interface is retained as an intramolecular interaction, and the novel α3/α4 interface has been added. The true dimer interface involving helices α3/α4 thus may not be required for the fundamental stabilization of the enzyme fold and may have evolved for other functional purposes. The product of apparent gene duplication, fusion, and further self-association is a large, cashew-shaped assembly rich in surface features such as projections, cavities, and charged and hydrophobic patches, representing potential interaction sites for other molecules. Particularly notable is the presence of a central cavity that is unique among Erv enzymes.
Although NCLDVs are morphologically diverse, they do share distantly related disulfide-bonded structural proteins that may be substrates of the viral sulfhydryl oxidases (16, 20). These proteins are associated with the viral outer membranes and have important roles in the virion assembly process. Examples include the L1R protein in poxviruses (36) and pE248R in ASFV (37). Baculoviruses are morphologically and compositionally distinct from NCLDVs, such that clear counterparts to the NCLDV structural proteins are not evident in baculoviruses (21). Therefore, it is difficult to propose putative sulfhydryl oxidase substrates in baculoviruses on the basis of information derived from NCLDVs. However, despite their distant relationship, common to both baculoviruses and NCLDVs is a severe drop in infectious virion production upon the elimination of sulfhydryl oxidase activity (25, 31, 39, 48), suggesting a fundamental role for disulfide formation in both cases. The molecular defects that lead to a dramatic drop in the number of infectious baculovirus particles produced in the absence of Ac92 have not yet been reported.
Baculoviruses generally are more complex than NCLDVs, with a biphasic infection cycle yielding two structurally distinct virion forms, the budded virions (BVs) and the occlusion-derived virions (ODVs). Common to both BVs and ODVs is the internal nucleocapsid, but the two forms have different outer envelope protein compositions, and the lipid membranes of the outer envelopes are likely to arise from different sources within the cell (42). Ac92 is produced from about 12 h postinfection (hpi) onwards (31, 48) and thus is present throughout the assembly of both the BV (12 to 24 hpi) and ODV (24 to 48 hpi) forms. Indeed, Ac92 has been observed to be physically associated with both BV and ODV particles (31, 48).
A number of possible functions for disulfide formation in the baculovirus infection cycle can be considered. Due to the widespread conservation of a sulfhydryl oxidase in baculoviruses and the demonstrated effect of Ac92 elimination on AcMNPV virion production and infectivity, one may postulate the existence of a general, essential substrate with a key role in virion assembly and/or infectivity. However, this general, conserved role does not rule out the participation of Ac92 in other minor or virus-specific processes. The conserved, essential substrate of Ac92 is not yet known, but the list may be shortened by the following considerations. The gp64 protein, which is the major outer envelope component of BVs, has both intramolecular and intersubunit disulfide bonds (23, 26). However, gp64 and other BV outer membrane proteins undergo oxidative folding in the endoplasmic reticulum and presumably use cellular systems for the introduction of disulfide bonds in this compartment (32). A similar argument can be used against the likelihood of a number of ODV envelope proteins being Ac92 substrates (5). Another possible target for baculovirus sulfhydryl oxidase activity is not a component of the virion itself: a fascinating phenomenon in baculovirus-infected cells is the formation of intracellular crystals, called polyhedra, that are capable of encapsulating entire ODVs. These crystals are composed of the protein polyhedrin, and disulfide bonds were observed linking polyhedrin molecules in crystals formed in vivo (8, 22). However, polyhedra form even in the absence of Ac92 (48), indicating either that disulfides are not essential for intracellular polyhedrin crystallization or that the formation of disulfides in polyhedra does not require Ac92. Presumably, the general Ac92 substrate is an essential factor widely present in BV forms of baculoviruses, does not traverse the secretory pathway, and contains cytosolic/nuclear domains requiring disulfide bonds for their folding and function.
Although the polyhedra themselves were not disrupted by the elimination of Ac92, a secondary role of Ac92 in ODV encapsulation nevertheless is possible, at least in alphabaculoviruses. Two striking features of the Ac92 knockout in AcMNPV were the lack of the self assembly of nucleocapsids to form bundles, as typically occurs before membrane encapsulation during ODV formation, and a decrease in incorporation into polyhedra of the atypical single-nucleocapsid ODVs that did form (48). These observations are consistent with the concentration of Ac92 enzyme in the ring zone of infected cells, where nucleocapsid self assembly and ODV formation and encapsulation into polyhedra occur (31). The possibility of a disulfide connection between the ODV envelope and the polyhedrin matrix is supported by the presence of an unpaired, conserved cysteine in a disordered peptide within polyhedra crystals (8). Furthermore, reducing agents are required to liberate the ODV envelope proteoglycan, PEP, from polyhedra (46). As the incorporation of ODVs into polyhedra begins to occur only at 24 hpi, putative roles for Ac92 in virion bundling and encapsulation would be in addition to important but as-yet unidentified functions during BV assembly at earlier times postinfection, as noted above.
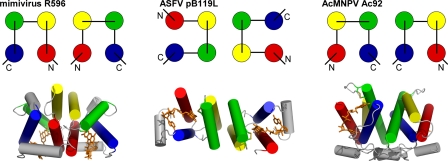
Only 10 core genes are shared among the diverse NCLDVs (49), and a few dozen are shared among baculoviruses (19). A much smaller subset of these, including Ac92, is shared across NCLDVs and baculoviruses. Catalyzed disulfide bond formation thus represents a crucial and widespread activity encoded within the genomes of numerous large double-stranded DNA viruses. It remains to be seen whether the targets of sulfhydryl oxidase activity in NCLDVs and baculoviruses are related, or whether related enzymes have been adopted for unrelated purposes in the two viral classes. In any case, the basic biochemical activity exhibited by Ac92 and its counterparts in NCLDVs is present in exceedingly varied structural contexts (Fig. 5), far surpassing the range of diversity observed for cellular enzymes of the same family.
Fig. 5.
Three viral sulfhydryl oxidases use three orthogonal interfaces for dimerization. A topology diagram demonstrating the helices buried in the interface of the FAD-binding, sulfhydryl oxidase domain for each viral enzyme dimer is shown above a representation of the dimer structure, with helices shown as cylinders. The four primary helices in each bundle are shown in red, yellow, green, and blue from amino to carboxy terminus. The ASFV structure is shown looking down the 2-fold axis of symmetry. The mimivirus and baculovirus structures are presented with the 2-fold axis vertical in the plane of the page.
ACKNOWLEDGMENTS
This work was supported by the Kimmelman Center for Macromolecular Assemblies.
We thank Emmanuel Levy and Nor Chejanovsky for helpful discussions.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 2. Adams P. D., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allen S., Balabanidou V., Sideris D. P., Lisowsky T., Tokatlidis K. 2005. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 353:937–944 [DOI] [PubMed] [Google Scholar]
- 4. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67:271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braunagel S. C., Summers M. D. 2007. Molecular biology of the baculovirus occlusion-derived virus envelope. Curr. Drug Targets 8:1084–1095 [DOI] [PubMed] [Google Scholar]
- 6. Brünger A. T., et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 7. Coppock D. L., Cina-Poppe D., Gilleran S. 1998. The quiescin Q6 gene (QSCN6) is a fusion of two ancient gene families: thioredoxin and ERV1. Genomics 54:460–468 [DOI] [PubMed] [Google Scholar]
- 8. Coulibaly F., et al. 2009. The atomic structure of baculovirus polyhedra reveals the independent emergence of infectious crystals in DNA and RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 106:22205–22210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daithankar V. N., Farrell S. R., Thorpe C. 2009. Augmenter of liver regeneration: substrate specificity of a flavin-dependent oxidoreductase from the mitochondrial intermembrane space. Biochemistry 48:4828–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daithankar V. N., Schaefer S. A., Dong M., Bahnson B. J., Thorpe C. 2010. Structure of the human sulfhydryl oxidase augmenter of liver regeneration and characterization of a human mutation causing an autosomal recessive myopathy. Biochemistry 49:6737–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Demeler B. 2005. UltraScan. A comprehensive data analysis software package for analytical ultracentrifugation experiments, p. 210–229In Scott D. J., Harding S. E., Rowe A. J.(ed.), Modern analytical ultracentrifugation: techniques and methods. Royal Society of Chemistry, London, United Kingdom [Google Scholar]
- 12. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 13. Fass D. 2008. The Erv family of sulfhydryl oxidases. Biochim. Biophys. Acta 1783:557–566 [DOI] [PubMed] [Google Scholar]
- 14. Gerber J., Muhlenhoff U., Hofhaus G., Lill R., Lisowsky T. 2001. Yeast ERV2p is the first microsomal FAD-linked sulfhydryl oxidase of the Erv1p/Alrp protein family. J. Biol. Chem. 276:23486–23491 [DOI] [PubMed] [Google Scholar]
- 15. Gross E., Sevier C. S., Vala A., Kaiser C. A., Fass D. 2002. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat. Struct. Biol. 9:61–67 [DOI] [PubMed] [Google Scholar]
- 16. Hakim M., Fass D. 2010. Cytosolic disulfide bond formation in cells infected with large nucleocytoplasmic DNA viruses. Antioxid. Redox Signal. 13:1261–1271 [DOI] [PubMed] [Google Scholar]
- 17. Hakim M., Fass D. 2009. Dimer interface migration in a viral sulfhydryl oxidase. J. Mol. Biol. 391:758–768 [DOI] [PubMed] [Google Scholar]
- 18. Heckler E. J., Alon A., Fass D., Thorpe C. 2008. Human quiescin-sulfhydryl oxidase, QSOX1: probing internal redox steps by mutagenesis. Biochemistry 47:4955–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herniou E. A., Olszewski J. A., Cory J. S., O'Reilly D. R. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211–234 [DOI] [PubMed] [Google Scholar]
- 20. Iyer L. M., Aravind L., Koonin E. V. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iyer L. M., Balaji S., Koonin E. V., Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156–184 [DOI] [PubMed] [Google Scholar]
- 22. Ji X., et al. 2010. How baculovirus polyhedra fit square pegs into round holes to robustly package viruses. EMBO J. 29:505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kadlec J., Loureiro S., Abrescia N. G., Stuart D. I., Jones I. M. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024–1030 [DOI] [PubMed] [Google Scholar]
- 24. Kodali V. K., Thorpe C. 2010. Oxidative protein folding and the Quiescin-sulfhydryl oxidase family of flavoproteins. Antioxid. Redox Signal. 13:1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lewis T., et al. 2000. An African swine fever virus ERV1-ALR homologue, 9GL, affects virion maturation and viral growth in macrophages and viral virulence in swine. J. Virol. 74:1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Z., Blissard G. W. 2010. Baculovirus GP64 disulfide bonds: the intermolecular disulfide bond of Autographa californica multicapsid nucleopolyhedrovirus GP64 is not essential for membrane fusion and virion budding. J. Virol. 84:8584–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lisowsky T. 1992. Dual function of a new nuclear gene for oxidative phosphorylation and vegetative growth in yeast. Mol. Gen. Genet. 232:58–64 [DOI] [PubMed] [Google Scholar]
- 28. Long C. M., Rohrmann G. F., Merrill G. F. 2009. The conserved baculovirus protein p33 (Ac92) is a flavin adenine dinucleotide-linked sulfhydryl oxidase. Virology 388:231–235 [DOI] [PubMed] [Google Scholar]
- 29. McCoy A. J., et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mesecke N., et al. 2005. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121:1059–1069 [DOI] [PubMed] [Google Scholar]
- 31. Nie Y., Fang M., Theilmann D. A. 2011. Autographa californica multiple nucleopolyhedrovirus core gene ac92 (p33) is required for efficient budded virus production. Virology 409:38–45 [DOI] [PubMed] [Google Scholar]
- 32. Oomens A. G., Monsma S. A., Blissard G. W. 1995. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology 209:592–603 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276:307–326 [DOI] [PubMed] [Google Scholar]
- 34. Pawlowski R., Jura J. 2006. ALR and liver regeneration. Mol. Cell. Biochem. 288:159–169 [DOI] [PubMed] [Google Scholar]
- 35. Raje S., Thorpe C. 2003. Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: role of a thioredoxin domain in disulfide bond formation. Biochemistry 42:4560–4568 [DOI] [PubMed] [Google Scholar]
- 36. Ravanello M. P., Hruby D. E. 1994. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly. J. Virol. 68:6401–6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodríguez I., Nogal M. L., Redrejo-Rodriguez M., Bustos M. J., Salas M. L. 2009. The African swine fever virus virion membrane protein pE248R is required for virus infectivity and an early postentry event. J. Virol. 83:12290–12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodríguez I., et al. 2006. African swine fever virus pB119L protein is a flavin adenine dinucleotide-linked sulfhydryl oxidase. J. Virol. 80:3157–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Senkevich T. G., Weisberg A. S., Moss B. 2000. Vaccinia virus E10R protein is associated with the membranes of intracellular mature virions and has a role in morphogenesis. Virology 278:244–252 [DOI] [PubMed] [Google Scholar]
- 40. Senkevich T. G., White C. L., Koonin E. V., Moss B. 2000. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. U. S. A. 97:12068–12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sevier C. S., Cuozzo J. W., Vala A., Aslund F., Kaiser C. A. 2001. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat. Cell Biol. 3:874–882 [DOI] [PubMed] [Google Scholar]
- 42. Slack J., Arif B. M. 2007. The baculoviruses occlusion-derived virus: virion structure and function. Adv. Virus Res. 69:99–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vala A., Sevier C. S., Kaiser C. A. 2005. Structural determinants of substrate access to the disulfide oxidase Erv2p. J. Mol. Biol. 354:952–966 [DOI] [PubMed] [Google Scholar]
- 44. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. 1993. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229:105–124 [DOI] [PubMed] [Google Scholar]
- 45. Vitu E., Bentzur M., Lisowsky T., Kaiser C. A., Fass D. 2006. Gain of function in an ERV/ALR sulfhydryl oxidase by molecular engineering of the shuttle disulfide. J. Mol. Biol. 362:89–101 [DOI] [PubMed] [Google Scholar]
- 46. Whitt M. A., Manning J. S. 1988. A phosphorylated 34-kDa protein and a subpopulation of polyhedrin are thiol linked to the carbohydrate layer surrounding a baculovirus occlusion body. Virology 163:33–42 [DOI] [PubMed] [Google Scholar]
- 47. Wu C. K., Dailey T. A., Dailey H. A., Wang B. C., Rose J. P. 2003. The crystal structure of augmenter of liver regeneration: a mammalian FAD-dependent sulfhydryl oxidase. Protein Sci. 12:1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu W., Passarelli A. L. 2010. Autographa californica multiple nucleopolyhedrovirus Ac92 (ORF92, P33) is required for budded virus production and multiply enveloped occlusion-derived virus formation. J. Virol. 84:12351–12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yutin N., Wolf Y. I., Raoult D., Koonin E. V. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 6:223. [DOI] [PMC free article] [PubMed] [Google Scholar]