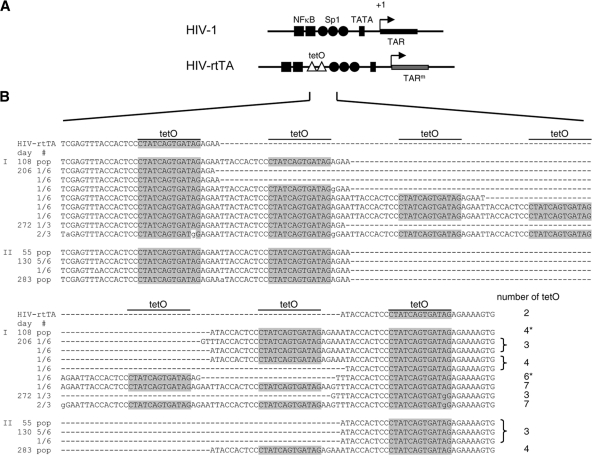
Fig. 2.
Evolution of HIV-rtTA-Tatstop. (A) Comparison of the U3-R regions from HIV-1 and HIV-rtTA. The HIV-rtTA variants have two tetO elements inserted between the NF-κB and Sp1 binding sites and several nucleotide substitutions in TAR (TARm). (B) Mutations observed in the tetO region of the HIV-rtTA-Tatstop mutant upon culturing with dox for up to 272 days (culture I) or 283 days (culture II). Cellular proviral DNA was isolated at 108, 206, and 272 days (culture I) and at 55, 130, and 283 days (culture II), and the LTR region was subsequently PCR amplified and either directly sequenced (population [pop] sequence; indicated on the left) or cloned into the TA-cloning vector followed by sequencing of three to six TA clones for each sample (with the frequency at which each sequence was observed indicated on the left). The nucleotide sequences of the tetO region between the NF-κB binding sites and the SpI binding sites are shown for HIV-rtTA-Tatstop (upper line) and the evolved viruses. The tetO core sequence is boxed in gray. The number of tetO elements present in each sequence is indicated on the right, with asterisks indicating the sequences that were recloned into HIV-rtTA-Tatstop (Fig. 3). Nucleotide substitutions are indicated with lowercase characters. Δ, nucleotide deletion.