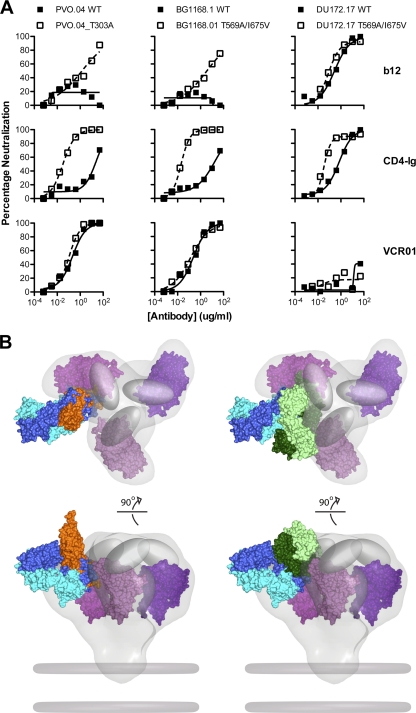
Fig. 3.
Impact of conformational constraints on neutralization by b12, CD4-Ig, and VRC01. (A) Env pseudoviruses PVO.04 (left) and BG1168.1 (middle) are relatively resistant to b12 and CD4-Ig. In the setting of the gp120 T303A mutation or the gp41 T569A/I675V mutations that result in a more open Env conformation, these viruses show enhanced neutralization sensitivity to b12 and CD4-Ig. In contrast, both the wild-type and mutant versions of these viruses were equally sensitive to VRC01-mediated neutralization. Virus DU172.17 (right) was resistant to VRC01, and this resistance was not affected by the Env mutations that alter conformation. (B) Model of VRC01, CD4, and b12 binding to the HIV-1 functional spike. Top (top) and front (bottom) views of the density map surface of the HIV-1 spike, presenting three gp120 core protomers in magenta (39) (Electron Microscopy Database [EMDB]: EMD-5019; Protein Data Bank [PDB]: 3DNN) and viral membrane in dark gray. To analyze the relative binding orientations of b12, CD4, and VRC01, the X-ray coordinates for gp120, in the b12, VRC01, and CD4/17b/bound conformations (PDB: 2NY7, 3NGB, and 2NY3), respectively, were superimposed on the same gp120 core protomer (PDB: 3DNN). The molecular surface of the b12-Fab fragment is green; the VRC01 Fab is blue. Within each color-coded Fab, the heavy chains are shaded darker than the light chains. The molecular surface of sCD4 is orange, and the variable loops are represented as gray ovals. Note that in this model, b12 and CD4 are more loop proximal than is VRC01.